A roadmap for interpreting (13)C metabolite labeling patterns from cells
- PMID: 25731751
- PMCID: PMC4552607
- DOI: 10.1016/j.copbio.2015.02.003
A roadmap for interpreting (13)C metabolite labeling patterns from cells
Abstract
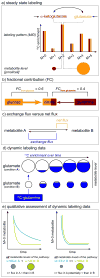
Measuring intracellular metabolism has increasingly led to important insights in biomedical research. (13)C tracer analysis, although less information-rich than quantitative (13)C flux analysis that requires computational data integration, has been established as a time-efficient method to unravel relative pathway activities, qualitative changes in pathway contributions, and nutrient contributions. Here, we review selected key issues in interpreting (13)C metabolite labeling patterns, with the goal of drawing accurate conclusions from steady state and dynamic stable isotopic tracer experiments.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

References
-
- Toya Y, Shimizu H. Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnology Advances. 2013;31:818–826. - PubMed
-
- Hiller K, Metallo C, Stephanopoulos G. Elucidation of Cellular Metabolism Via Metabolomics and Stable-Isotope Assisted Metabolomics. Current Pharmaceutical Biotechnology. 2011:12. - PubMed
-
- Hiller K, Metallo CM. Profiling metabolic networks to study cancer metabolism. Current Opinion in Biotechnology. 2013;24:60–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12022/6/MRC_/Medical Research Council/United Kingdom
- 21140/CRUK_/Cancer Research UK/United Kingdom
- R01 CA157996/CA/NCI NIH HHS/United States
- R01 DK078184/DK/NIDDK NIH HHS/United States
- R01 AI081773/AI/NIAID NIH HHS/United States
- R01 DK092606/DK/NIDDK NIH HHS/United States
- R01 CA160458/CA/NCI NIH HHS/United States
- 18278/CRUK_/Cancer Research UK/United Kingdom
- 19702/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_1101/3/MRC_/Medical Research Council/United Kingdom
- R56 AI081773/AI/NIAID NIH HHS/United States
- S10 RR029119/RR/NCRR NIH HHS/United States
- 17728/CRUK_/Cancer Research UK/United Kingdom
- R00 CA168997/CA/NCI NIH HHS/United States
- 18274/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

