Interaction with OGG1 is required for efficient recruitment of XRCC1 to base excision repair and maintenance of genetic stability after exposure to oxidative stress
- PMID: 25733688
- PMCID: PMC4387214
- DOI: 10.1128/MCB.00134-15
Interaction with OGG1 is required for efficient recruitment of XRCC1 to base excision repair and maintenance of genetic stability after exposure to oxidative stress
Abstract
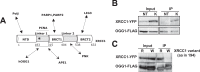
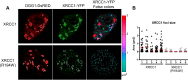
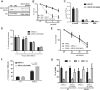
XRCC1 is an essential protein required for the maintenance of genomic stability through its implication in DNA repair. The main function of XRCC1 is associated with its role in the single-strand break (SSB) and base excision repair (BER) pathways that share several enzymatic steps. We show here that the polymorphic XRCC1 variant R194W presents a defect in its interaction with the DNA glycosylase OGG1 after oxidative stress. While proficient for single-strand break repair (SSBR), this variant does not colocalize with OGG1, reflecting a defect in its involvement in BER. Consistent with a role of XRCC1 in the coordination of the BER pathway, induction of oxidative base damage in XRCC1-deficient cells complemented with the R194W variant results in increased genetic instability as revealed by the accumulation of micronuclei. These data identify a specific molecular role for the XRCC1-OGG1 interaction in BER and provide a model for the effects of the R194W variant identified in molecular cancer epidemiology studies.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
