Promyelocytic Leukemia Protein Isoform II Promotes Transcription Factor Recruitment To Activate Interferon Beta and Interferon-Responsive Gene Expression
- PMID: 25733689
- PMCID: PMC4405644
- DOI: 10.1128/MCB.01478-14
Promyelocytic Leukemia Protein Isoform II Promotes Transcription Factor Recruitment To Activate Interferon Beta and Interferon-Responsive Gene Expression
Abstract
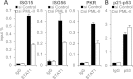
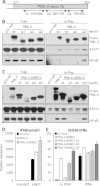
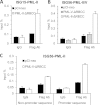
To trigger type I interferon (IFN) responses, pattern recognition receptors activate signaling cascades that lead to transcription of IFN and IFN-stimulated genes (ISGs). The promyelocytic leukemia (PML) protein has been implicated in these responses, although its role has not been defined. Here, we show that PML isoform II (PML-II) is specifically required for efficient induction of IFN-β transcription and of numerous ISGs, acting at the point of transcriptional complex assembly on target gene promoters. PML-II associated with specific transcription factors NF-κB and STAT1, as well as the coactivator CREB-binding protein (CBP), to facilitate transcriptional complex formation. The absence of PML-II substantially reduced binding of these factors and IFN regulatory factor 3 (IRF3) to IFN-β or ISGs promoters and sharply reduced gene activation. The unique C-terminal domain of PML-II was essential for its activity, while the N-terminal RBCC motif common to all PML isoforms was dispensable. We propose a model in which PML-II contributes to the transcription of multiple genes via the association of its C-terminal domain with relevant transcription complexes, which promotes the stable assembly of these complexes at promoters/enhancers of target genes, and that in this way PML-II plays a significant role in the development of type I IFN responses.
Copyright © 2015, Chen et al.
Figures
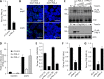
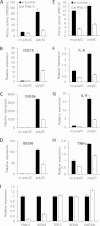


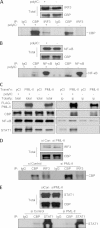
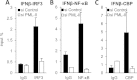
Similar articles
-
PML positively regulates interferon gamma signaling.Biochimie. 2011 Mar;93(3):389-98. doi: 10.1016/j.biochi.2010.11.005. Epub 2010 Nov 27. Biochimie. 2011. PMID: 21115099
-
Role of promyelocytic leukemia protein in host antiviral defense.J Interferon Cytokine Res. 2011 Jan;31(1):145-58. doi: 10.1089/jir.2010.0111. Epub 2011 Jan 3. J Interferon Cytokine Res. 2011. PMID: 21198351 Review.
-
Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus.PLoS Pathog. 2015 Mar 26;11(3):e1004785. doi: 10.1371/journal.ppat.1004785. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25812002 Free PMC article.
-
Direct transcriptional activation of promyelocytic leukemia protein by IFN regulatory factor 3 induces the p53-dependent growth inhibition of cancer cells.Cancer Res. 2007 Dec 1;67(23):11133-40. doi: 10.1158/0008-5472.CAN-07-1342. Cancer Res. 2007. PMID: 18056437
-
[Implication of PML nuclear bodies in intrinsic and innate immunity].Med Sci (Paris). 2014 Aug-Sep;30(8-9):765-71. doi: 10.1051/medsci/20143008014. Epub 2014 Sep 1. Med Sci (Paris). 2014. PMID: 25174753 Review. French.
Cited by
-
PML-NB-dependent type I interferon memory results in a restricted form of HSV latency.EMBO Rep. 2021 Sep 6;22(9):e52547. doi: 10.15252/embr.202152547. Epub 2021 Jul 1. EMBO Rep. 2021. PMID: 34197022 Free PMC article.
-
The essential role of guinea pig cytomegalovirus (GPCMV) IE1 and IE2 homologs in viral replication and IE1-mediated ND10 targeting.Virology. 2017 Apr;504:122-140. doi: 10.1016/j.virol.2017.01.023. Epub 2017 Feb 10. Virology. 2017. PMID: 28189970 Free PMC article.
-
The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection.Virus Res. 2020 Aug;285:198015. doi: 10.1016/j.virusres.2020.198015. Epub 2020 May 13. Virus Res. 2020. PMID: 32416261 Free PMC article. Review.
-
Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1.PLoS Pathog. 2020 May 4;16(5):e1008537. doi: 10.1371/journal.ppat.1008537. eCollection 2020 May. PLoS Pathog. 2020. PMID: 32365141 Free PMC article.
-
PML-II regulates ERK and AKT signal activation and IFNα-induced cell death.Cell Commun Signal. 2021 Jul 2;19(1):70. doi: 10.1186/s12964-021-00756-5. Cell Commun Signal. 2021. PMID: 34215258 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
