Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells
- PMID: 25733697
- PMCID: PMC4433415
- DOI: 10.1158/1541-7786.MCR-14-0412
Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells
Abstract
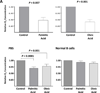
While reviewing chronic lymphocytic leukemia (CLL) bone marrow slides, we identified cytoplasmic lipid vacuoles in CLL cells but not in normal B cells. Because lipoprotein lipase (LPL), which catalyzes hydrolysis of triglycerides into free fatty acids (FFA), is aberrantly expressed in CLL, we investigated whether LPL regulates the oxidative metabolic capacity of CLL cells. We found that unlike normal B cells, CLL cells metabolize FFAs. Because STAT3 is constitutively activated in CLL cells and because we identified putative STAT3 binding sites in the LPL promoter, we sought to determine whether STAT3 drives the aberrant expression of LPL. Transfection of luciferase reporter gene constructs driven by LPL promoter fragments into MM1 cells revealed that STAT3 activates the LPL promoter. In addition, chromatin immunoprecipitation confirmed that STAT3 binds to the LPL promoter. Furthermore, transfection of CLL cells with STAT3-shRNA downregulated LPL transcripts and protein levels, confirming that STAT3 activates the LPL gene. Finally, transfection of CLL cells with LPL-siRNAs decreased the capacity of CLL cells to oxidize FFAs and reduced cell viability.
Implications: Our study suggests that CLL cells adopt their metabolism to oxidize FFA. Activated STAT3 induces LPL, which catalyzes the hydrolysis of triglycerides into FFA. Therefore, inhibition of STAT3 is likely to prevent the capacity of CLL cells to utilize FFA.
©2015 American Association for Cancer Research.
Conflict of interest statement
The authors declare no competing financial interests.
Figures









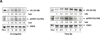
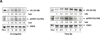
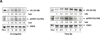
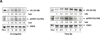
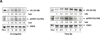



References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Damle RN, Ghiotto F, Valetto A, et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood. 2002;99(11):4087–4093. - PubMed
-
- Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol. 2003;170(2):686–694. - PubMed
-
- Chiorazzi N. Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best Pract Res Clin Haematol. 2007;20(3):399–413. - PubMed

