Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells
- PMID: 25733714
- PMCID: PMC4347635
- DOI: 10.1083/jcb.201406099
Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells
Abstract
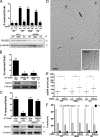
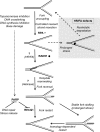
Replication fork reversal protects forks from breakage after poisoning of Topoisomerase 1. We here investigated fork progression and chromosomal breakage in human cells in response to a panel of sublethal genotoxic treatments, using other topoisomerase poisons, DNA synthesis inhibitors, interstrand cross-linking inducers, and base-damaging agents. We used electron microscopy to visualize fork architecture under these conditions and analyzed the association of specific molecular features with checkpoint activation. Our data identify replication fork uncoupling and reversal as global responses to genotoxic treatments. Both events are frequent even after mild treatments that do not affect fork integrity, nor activate checkpoints. Fork reversal was found to be dependent on the central homologous recombination factor RAD51, which is consistently present at replication forks independently of their breakage, and to be antagonized by poly (ADP-ribose) polymerase/RECQ1-regulated restart. Our work establishes remodeling of uncoupled forks as a pivotal RAD51-regulated response to genotoxic stress in human cells and as a promising target to potentiate cancer chemotherapy.
© 2015 Zellweger et al.
Figures

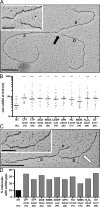
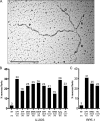

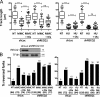
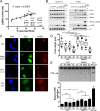
References
-
- Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., and Groth A.. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16:281–293 10.1038/ncb2918 - DOI - PMC - PubMed
-
- Bermejo R., Capra T., Jossen R., Colosio A., Frattini C., Carotenuto W., Cocito A., Doksani Y., Klein H., Gómez-González B., et al. . 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 146:233–246 10.1016/j.cell.2011.06.033 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

