Evaluating methods for modeling epistasis networks with application to head and neck cancer
- PMID: 25733798
- PMCID: PMC4332043
- DOI: 10.4137/CIN.S17289
Evaluating methods for modeling epistasis networks with application to head and neck cancer
Abstract
Epistasis helps to explain how multiple single-nucleotide polymorphisms (SNPs) interact to cause disease. A variety of tools have been developed to detect epistasis. In this article, we explore the strengths and weaknesses of an information theory approach for detecting epistasis and compare it to the logistic regression approach through simulations. We consider several scenarios to simulate the involvement of SNPs in an epistasis network with respect to linkage disequilibrium patterns among them and the presence or absence of main and interaction effects. We conclude that the information theory approach more efficiently detects interaction effects when main effects are absent, whereas, in general, the logistic regression approach is appropriate in all scenarios but results in higher false positives. We compute epistasis networks for SNPs in the FSD1L gene using a two-phase head and neck cancer genome-wide association study involving 2,185 cases and 4,507 controls to demonstrate the practical application of the methods.
Keywords: epistasis; head and neck cancer; information theory; networks; regression.
Figures
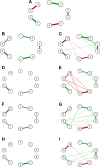
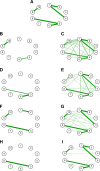



References
-
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17(R2):R109–15. - PubMed
-
- Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11(20):2463–8. - PubMed
-
- Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34(2):88–95. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

