TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis
- PMID: 25733846
- PMCID: PMC4386332
- DOI: 10.1073/pnas.1421587112
TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis
Abstract
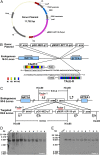
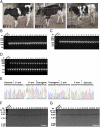
Transcription activator-like effector nuclease (TALEN)-mediated genome modification has been applied successfully to create transgenic animals in various species, such as mouse, pig, and even monkey. However, transgenic cattle with gene knockin have yet to be created using TALENs. Here, we report site-specific knockin of the transcription activator-like effector (TALE) nickase-mediated SP110 nuclear body protein gene (SP110) via homologous recombination to produce tuberculosis-resistant cattle. In vitro and in vivo challenge and transmission experiments proved that the transgenic cattle are able to control the growth and multiplication of Mycobacterium bovis, turn on the apoptotic pathway of cell death instead of necrosis after infection, and efficiently resist the low dose of M. bovis transmitted from tuberculous cattle in nature. In this study, we developed TALE nickases to modify the genome of Holstein-Friesian cattle, thereby engineering a heritable genome modification that facilitates resistance to tuberculosis.
Keywords: TALEN; disease resistance; homologous recombination; single-strand break; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Tuberculosis-resistant transgenic cattle.Proc Natl Acad Sci U S A. 2015 Mar 31;112(13):3854-5. doi: 10.1073/pnas.1502972112. Epub 2015 Mar 17. Proc Natl Acad Sci U S A. 2015. PMID: 25829535 Free PMC article. No abstract available.
-
Equation-free modeling unravels the behavior of complex ecological systems.Proc Natl Acad Sci U S A. 2015 Mar 31;112(13):3856-7. doi: 10.1073/pnas.1503154112. Epub 2015 Mar 17. Proc Natl Acad Sci U S A. 2015. PMID: 25829536 Free PMC article. No abstract available.
References
-
- Sommer D, et al. Efficient genome engineering by targeted homologous recombination in mouse embryos using transcription activator-like effector nucleases. Nat Commun. 2014;5:3045. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

