Discovery of novel new Delhi metallo-β-lactamases-1 inhibitors by multistep virtual screening
- PMID: 25734558
- PMCID: PMC4348537
- DOI: 10.1371/journal.pone.0118290
Discovery of novel new Delhi metallo-β-lactamases-1 inhibitors by multistep virtual screening
Abstract
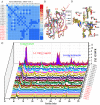
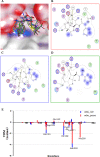
The emergence of NDM-1 containing multi-antibiotic resistant "Superbugs" necessitates the needs of developing of novel NDM-1inhibitors. In this study, we report the discovery of novel NDM-1 inhibitors by multi-step virtual screening. From a 2,800,000 virtual drug-like compound library selected from the ZINC database, we generated a focused NDM-1 inhibitor library containing 298 compounds of which 44 chemical compounds were purchased and evaluated experimentally for their ability to inhibit NDM-1 in vitro. Three novel NDM-1 inhibitors with micromolar IC50 values were validated. The most potent inhibitor, VNI-41, inhibited NDM-1 with an IC50 of 29.6 ± 1.3 μM. Molecular dynamic simulation revealed that VNI-41 interacted extensively with the active site. In particular, the sulfonamide group of VNI-41 interacts directly with the metal ion Zn1 that is critical for the catalysis. These results demonstrate the feasibility of applying virtual screening methodologies in identifying novel inhibitors for NDM-1, a metallo-β-lactamase with a malleable active site and provide a mechanism base for rational design of NDM-1 inhibitors using sulfonamide as a functional scaffold.
Conflict of interest statement
Figures





Similar articles
-
High-Throughput Virtual Screening, Molecular Dynamics Simulation, and Enzyme Kinetics Identified ZINC84525623 as a Potential Inhibitor of NDM-1.Int J Mol Sci. 2019 Feb 14;20(4):819. doi: 10.3390/ijms20040819. Int J Mol Sci. 2019. PMID: 30769822 Free PMC article.
-
Recent research and development of NDM-1 inhibitors.Eur J Med Chem. 2021 Nov 5;223:113667. doi: 10.1016/j.ejmech.2021.113667. Epub 2021 Jun 24. Eur J Med Chem. 2021. PMID: 34225181 Review.
-
Risedronate and Methotrexate Are High-Affinity Inhibitors of New Delhi Metallo-β-Lactamase-1 (NDM-1): A Drug Repurposing Approach.Molecules. 2022 Feb 14;27(4):1283. doi: 10.3390/molecules27041283. Molecules. 2022. PMID: 35209073 Free PMC article.
-
Diaryl-substituted azolylthioacetamides: Inhibitor discovery of New Delhi metallo-β-lactamase-1 (NDM-1).ChemMedChem. 2014 Nov;9(11):2445-8. doi: 10.1002/cmdc.201402249. Epub 2014 Jul 22. ChemMedChem. 2014. PMID: 25048031
-
New Delhi metallo-β-lactamase-1: structure, inhibitors and detection of producers.Future Med Chem. 2016 Jun;8(9):993-1012. doi: 10.4155/fmc-2016-0015. Epub 2016 Jun 2. Future Med Chem. 2016. PMID: 27253479 Review.
Cited by
-
Current Strategy for Targeting Metallo-β-Lactamase with Metal-Ion-Binding Inhibitors.Molecules. 2024 Aug 21;29(16):3944. doi: 10.3390/molecules29163944. Molecules. 2024. PMID: 39203022 Free PMC article. Review.
-
Identification of a Potential Inhibitor (MCULE-8777613195-0-12) of New Delhi Metallo-β-Lactamase-1 (NDM-1) Using In Silico and In Vitro Approaches.Molecules. 2022 Sep 13;27(18):5930. doi: 10.3390/molecules27185930. Molecules. 2022. PMID: 36144666 Free PMC article.
-
NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings.Clin Microbiol Rev. 2019 Jan 30;32(2):e00115-18. doi: 10.1128/CMR.00115-18. Print 2019 Mar 20. Clin Microbiol Rev. 2019. PMID: 30700432 Free PMC article. Review.
-
Machine Learning Models Identify Inhibitors of New Delhi Metallo-β-lactamase.J Chem Inf Model. 2024 May 27;64(10):3977-3991. doi: 10.1021/acs.jcim.3c02015. Epub 2024 May 10. J Chem Inf Model. 2024. PMID: 38727192 Free PMC article.
-
Virtual Screening and Experimental Testing of B1 Metallo-β-lactamase Inhibitors.J Chem Inf Model. 2018 Sep 24;58(9):1902-1914. doi: 10.1021/acs.jcim.8b00133. Epub 2018 Aug 29. J Chem Inf Model. 2018. PMID: 30107123 Free PMC article.
References
-
- Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005; 105: 395–424. - PubMed
-
- U.S. Centers for Disease Control and Prevention. Antibiotic resistance threats in the united states. Available: http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013.... Accessed 2013 Apr 23.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

