Comparative analysis of metagenomes from three methanogenic hydrocarbon-degrading enrichment cultures with 41 environmental samples
- PMID: 25734684
- PMCID: PMC4542035
- DOI: 10.1038/ismej.2015.22
Comparative analysis of metagenomes from three methanogenic hydrocarbon-degrading enrichment cultures with 41 environmental samples
Abstract
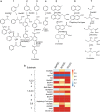
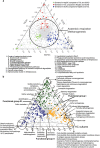
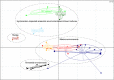
Methanogenic hydrocarbon metabolism is a key process in subsurface oil reservoirs and hydrocarbon-contaminated environments and thus warrants greater understanding to improve current technologies for fossil fuel extraction and bioremediation. In this study, three hydrocarbon-degrading methanogenic cultures established from two geographically distinct environments and incubated with different hydrocarbon substrates (added as single hydrocarbons or as mixtures) were subjected to metagenomic and 16S rRNA gene pyrosequencing to test whether these differences affect the genetic potential and composition of the communities. Enrichment of different putative hydrocarbon-degrading bacteria in each culture appeared to be substrate dependent, though all cultures contained both acetate- and H2-utilizing methanogens. Despite differing hydrocarbon substrates and inoculum sources, all three cultures harbored genes for hydrocarbon activation by fumarate addition (bssA, assA, nmsA) and carboxylation (abcA, ancA), along with those for associated downstream pathways (bbs, bcr, bam), though the cultures incubated with hydrocarbon mixtures contained a broader diversity of fumarate addition genes. A comparative metagenomic analysis of the three cultures showed that they were functionally redundant despite their enrichment backgrounds, sharing multiple features associated with syntrophic hydrocarbon conversion to methane. In addition, a comparative analysis of the culture metagenomes with those of 41 environmental samples (containing varying proportions of methanogens) showed that the three cultures were functionally most similar to each other but distinct from other environments, including hydrocarbon-impacted environments (for example, oil sands tailings ponds and oil-affected marine sediments). This study provides a basis for understanding key functions and environmental selection in methanogenic hydrocarbon-associated communities.
Figures


References
-
- Abu Laban N, Selesi D, Jobelius C, Meckenstock RU. Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol Ecol. 2009;68:300–311. - PubMed
-
- Abu Laban N, Selesi D, Rattei T, Tischler P, Meckenstock RU. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ Microbiol. 2010;12:2783–2796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

