Structural and functional investigation of flavin binding center of the NqrC subunit of sodium-translocating NADH:quinone oxidoreductase from Vibrio harveyi
- PMID: 25734798
- PMCID: PMC4348036
- DOI: 10.1371/journal.pone.0118548
Structural and functional investigation of flavin binding center of the NqrC subunit of sodium-translocating NADH:quinone oxidoreductase from Vibrio harveyi
Abstract
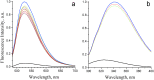
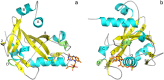
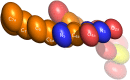
Na+-translocating NADH:quinone oxidoreductase (NQR) is a redox-driven sodium pump operating in the respiratory chain of various bacteria, including pathogenic species. The enzyme has a unique set of redox active prosthetic groups, which includes two covalently bound flavin mononucleotide (FMN) residues attached to threonine residues in subunits NqrB and NqrC. The reason of FMN covalent bonding in the subunits has not been established yet. In the current work, binding of free FMN to the apo-form of NqrC from Vibrio harveyi was studied showing very low affinity of NqrC to FMN in the absence of its covalent bonding. To study structural aspects of flavin binding in NqrC, its holo-form was crystallized and its 3D structure was solved at 1.56 Å resolution. It was found that the isoalloxazine moiety of the FMN residue is buried in a hydrophobic cavity and that its pyrimidine ring is squeezed between hydrophobic amino acid residues while its benzene ring is extended from the protein surroundings. This structure of the flavin-binding pocket appears to provide flexibility of the benzene ring, which can help the FMN residue to take the bended conformation and thus to stabilize the one-electron reduced form of the prosthetic group. These properties may also lead to relatively weak noncovalent binding of the flavin. This fact along with periplasmic location of the FMN-binding domains in the vast majority of NqrC-like proteins may explain the necessity of the covalent bonding of this prosthetic group to prevent its loss to the external medium.
Conflict of interest statement
Figures


References
-
- Rich PR, Meunier B, Ward FB. Predicted structure and possible ionmotive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus . FEBS Lett. 1995;375: 5–10. - PubMed
-
- Hayashi M, Hirai K, Unemoto T. Cloning of the Na+-translocating NADH-quinone reductase gene from the marine bacterium Vibrio alginolyticus and the expression of the beta-subunit in Escherichia coli . FEBS Lett. 1994;356: 330–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

