Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis
- PMID: 25734800
- PMCID: PMC4348521
- DOI: 10.1371/journal.pone.0117403
Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis
Abstract
Objective: Determine whether MS-specific DNA methylation profiles can be identified in whole blood or purified immune cells from untreated MS patients.
Methods: Whole blood, CD4+ and CD8+ T cell DNA from 16 female, treatment naïve MS patients and 14 matched controls was profiled using the HumanMethylation450K BeadChip. Genotype data were used to assess genetic homogeneity of our sample and to exclude potential SNP-induced DNA methylation measurement errors.
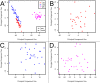
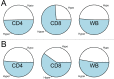
Results: As expected, significant differences between CD4+ T cells, CD8+ T cells and whole blood DNA methylation profiles were observed, regardless of disease status. Strong evidence for hypermethylation of CD8+ T cell, but not CD4+ T cell or whole blood DNA in MS patients compared to controls was observed. Genome-wide significant individual CpG-site DNA methylation differences were not identified. Furthermore, significant differences in gene DNA methylation of 148 established MS-associated risk genes were not observed.
Conclusion: While genome-wide significant DNA methylation differences were not detected for individual CpG-sites, strong evidence for DNA hypermethylation of CD8+ T cells for MS patients was observed, indicating a role for DNA methylation in MS. Further, our results suggest that large DNA methylation differences for CpG-sites tested here do not contribute to MS susceptibility. In particular, large DNA methylation differences for CpG-sites within 148 established MS candidate genes tested in our study cannot explain missing heritability. Larger studies of homogenous MS patients and matched controls are warranted to further elucidate the impact of CD8+ T cell and more subtle DNA methylation changes in MS development and pathogenesis.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

