Membrane-assisted growth of DNA origami nanostructure arrays
- PMID: 25734977
- PMCID: PMC4415451
- DOI: 10.1021/acsnano.5b00161
Membrane-assisted growth of DNA origami nanostructure arrays
Abstract
Biological membranes fulfill many important tasks within living organisms. In addition to separating cellular volumes, membranes confine the space available to membrane-associated proteins to two dimensions (2D), which greatly increases their probability to interact with each other and assemble into multiprotein complexes. We here employed two DNA origami structures functionalized with cholesterol moieties as membrane anchors--a three-layered rectangular block and a Y-shaped DNA structure--to mimic membrane-assisted assembly into hierarchical superstructures on supported lipid bilayers and small unilamellar vesicles. As designed, the DNA constructs adhered to the lipid bilayers mediated by the cholesterol anchors and diffused freely in 2D with diffusion coefficients depending on their size and number of cholesterol modifications. Different sets of multimerization oligonucleotides added to bilayer-bound origami block structures induced the growth of either linear polymers or two-dimensional lattices on the membrane. Y-shaped DNA origami structures associated into triskelion homotrimers and further assembled into weakly ordered arrays of hexagons and pentagons, which resembled the geometry of clathrin-coated pits. Our results demonstrate the potential to realize artificial self-assembling systems that mimic the hierarchical formation of polyhedral lattices on cytoplasmic membranes.
Keywords: DNA nanotechnology; DNA origami; arrays; cholesterol; clathrin; diffusion; lipid membrane.
Figures

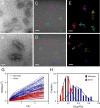
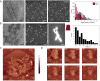

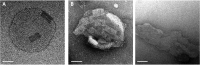
Comment in
-
Mimicking membrane-related biological events by DNA origami nanotechnology.ACS Nano. 2015 Apr 28;9(4):3418-20. doi: 10.1021/acsnano.5b01723. Epub 2015 Apr 16. ACS Nano. 2015. PMID: 25880224
References
-
- Axelrod D. Lateral Motion of Membrane Proteins and Biological Function. J. Membr. Biol. 1983, 75, 1–10. - PubMed
-
- Heldin C. H. Dimerization of Cell Surface Receptors in Signal Transduction. Cell 1995, 80, 213–223. - PubMed
-
- Doherty G. J.; McMahon H. T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

