IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics
- PMID: 25735289
- PMCID: PMC5148126
- DOI: 10.1118/1.4908210
IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics
Abstract
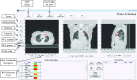
Purpose: Radiomics, which is the high-throughput extraction and analysis of quantitative image features, has been shown to have considerable potential to quantify the tumor phenotype. However, at present, a lack of software infrastructure has impeded the development of radiomics and its applications. Therefore, the authors developed the imaging biomarker explorer (IBEX), an open infrastructure software platform that flexibly supports common radiomics workflow tasks such as multimodality image data import and review, development of feature extraction algorithms, model validation, and consistent data sharing among multiple institutions.
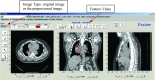
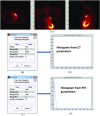
Methods: The IBEX software package was developed using the MATLAB and c/c++ programming languages. The software architecture deploys the modern model-view-controller, unit testing, and function handle programming concepts to isolate each quantitative imaging analysis task, to validate if their relevant data and algorithms are fit for use, and to plug in new modules. On one hand, IBEX is self-contained and ready to use: it has implemented common data importers, common image filters, and common feature extraction algorithms. On the other hand, IBEX provides an integrated development environment on top of MATLAB and c/c++, so users are not limited to its built-in functions. In the IBEX developer studio, users can plug in, debug, and test new algorithms, extending IBEX's functionality. IBEX also supports quality assurance for data and feature algorithms: image data, regions of interest, and feature algorithm-related data can be reviewed, validated, and/or modified. More importantly, two key elements in collaborative workflows, the consistency of data sharing and the reproducibility of calculation result, are embedded in the IBEX workflow: image data, feature algorithms, and model validation including newly developed ones from different users can be easily and consistently shared so that results can be more easily reproduced between institutions.
Results: Researchers with a variety of technical skill levels, including radiation oncologists, physicists, and computer scientists, have found the IBEX software to be intuitive, powerful, and easy to use. IBEX can be run at any computer with the windows operating system and 1GB RAM. The authors fully validated the implementation of all importers, preprocessing algorithms, and feature extraction algorithms. Windows version 1.0 beta of stand-alone IBEX and IBEX's source code can be downloaded.
Conclusions: The authors successfully implemented IBEX, an open infrastructure software platform that streamlines common radiomics workflow tasks. Its transparency, flexibility, and portability can greatly accelerate the pace of radiomics research and pave the way toward successful clinical translation.
Figures






References
-
- Chen H. Y., Yu S. L., Chen C. H., Chang G. C., Chen C. Y., Yuan A., Cheng C. L., Wang C. H., Terng H. J., Kao S. F., Chan W. K., Li H. N., Liu C. C., Singh S., Chen W. J., Chen J. J., and Yang P. C., “A five-gene signature and clinical outcome in non-small-cell lung cancer,” N. Engl. J. Med. 356, 11–20 (2007).10.1056/NEJMoa060096 - DOI - PubMed
-
- Eisenhauer E. A., Therasse P., Bogaerts J., Schwartz L. H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., and Verweij J., “New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1),” Eur. J. Cancer 45, 228–247 (2009).10.1016/j.ejca.2008.10.026 - DOI - PubMed
-
- Machtay M., Duan F., Siegel B. A., Snyder B. S., Gorelick J. J., Reddin J. S., Munden R., Johnson D. W., Wilf L. H., DeNittis A., Sherwin N., Cho K. H., Kim S. K., Videtic G., Neumann D. R., Komaki R., Macapinlac H., Bradley J. D., and Alavi A., “Prediction of survival by [18F]fluorodeoxyglucose positron emission tomography in patients with locally advanced non-small-cell lung cancer undergoing definitive chemoradiation therapy: Results of the ACRIN 6668/RTOG 0235 trial,” J. Clin. Oncol. 31, 3823–3830 (2013).10.1200/JCO.2012.47.5947 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

