Estimating the proportion of true null hypotheses when the statistics are discrete
- PMID: 25735771
- PMCID: PMC4495288
- DOI: 10.1093/bioinformatics/btv104
Estimating the proportion of true null hypotheses when the statistics are discrete
Abstract
Motivation: In high-dimensional testing problems π0, the proportion of null hypotheses that are true is an important parameter. For discrete test statistics, the P values come from a discrete distribution with finite support and the null distribution may depend on an ancillary statistic such as a table margin that varies among the test statistics. Methods for estimating π0 developed for continuous test statistics, which depend on a uniform or identical null distribution of P values, may not perform well when applied to discrete testing problems.
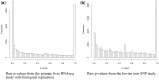
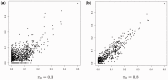
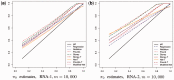
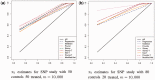
Results: This article introduces a number of π0 estimators, the regression and 'T' methods that perform well with discrete test statistics and also assesses how well methods developed for or adapted from continuous tests perform with discrete tests. We demonstrate the usefulness of these estimators in the analysis of high-throughput biological RNA-seq and single-nucleotide polymorphism data.
Availability and implementation: implemented in R.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Bancroft T., et al. (2013) Estimation of false discovery rate using sequential permutation p-values. Biometrics , 69, 1–7. - PubMed
-
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B , 57, 289–300.
-
- Benjamini Y., Hochberg Y. (2000) On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Behav. Educ. Stat. , 25, 60–83.
-
- Black M.A. (2004) A note on the adaptive control of false discovery rates. J. R. Stat. Soc. B , 66, 297–304.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

