Proteomic analysis of lettuce seed germination and thermoinhibition by sampling of individual seeds at germination and removal of storage proteins by polyethylene glycol fractionation
- PMID: 25736209
- PMCID: PMC4378177
- DOI: 10.1104/pp.15.00045
Proteomic analysis of lettuce seed germination and thermoinhibition by sampling of individual seeds at germination and removal of storage proteins by polyethylene glycol fractionation
Abstract
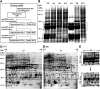
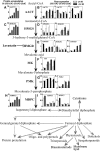
Germination and thermoinhibition in lettuce (Lactuca sativa 'Jianyexianfeng No. 1') seeds were investigated by a proteomic comparison among dry seeds, germinated seeds at 15°C, at 15°C after imbibition at 25°C for 48 h, or at 25°C in KNO3 (all sampled individually at germination), and ungerminated seeds at 25°C, a thermoinhibitory temperature. Before two-dimensional gel electrophoresis analysis, storage proteins (greater than 50% of total extractable protein) were removed by polyethylene glycol precipitation, which significantly improved the detection of less abundant proteins on two-dimensional gels. A total of 108 protein spots were identified to change more than 2-fold (P<0.05) in abundance in at least one germination treatment. Nineteen proteins increasing and one protein decreasing in abundance during germination had higher abundance in germinated 15°C, 15°C after imbibition at 25°C for 48 h, and 25°C in KNO3 seeds than in ungerminated 25°C seeds. Gene expression of 12 of those proteins correlated well with the protein accumulation. Methionine metabolism, ethylene production, lipid mobilization, cell elongation, and detoxification of aldehydes were revealed to be potentially related to lettuce seed germination and thermoinhibition. Accumulation of three proteins and expression of five genes participating in the mevalonate (MVA) pathway of isoprenoid biosynthesis correlated positively with seed germinability. Inhibition of this pathway by lovastatin delayed seed germination and increased the sensitivity of germination to abscisic acid. MVA pathway-derived products, cytokinins, partially reversed the lovastatin inhibition of germination and released seed thermoinhibition at 25°C. We conclude that the MVA pathway for isoprenoid biosynthesis is involved in lettuce seed germination and thermoinhibition.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures




Similar articles
-
Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic Acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes.Plant Physiol. 2008 Oct;148(2):926-47. doi: 10.1104/pp.108.125807. Epub 2008 Aug 27. Plant Physiol. 2008. PMID: 18753282 Free PMC article.
-
Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance.Plant Cell. 2013 Mar;25(3):884-900. doi: 10.1105/tpc.112.108902. Epub 2013 Mar 15. Plant Cell. 2013. PMID: 23503626 Free PMC article.
-
Genetic Variation for Thermotolerance in Lettuce Seed Germination Is Associated with Temperature-Sensitive Regulation of ETHYLENE RESPONSE FACTOR1 (ERF1).Plant Physiol. 2016 Jan;170(1):472-88. doi: 10.1104/pp.15.01251. Epub 2015 Nov 16. Plant Physiol. 2016. PMID: 26574598 Free PMC article.
-
Advance in the Thermoinhibition of Lettuce (Lactuca sativa L.) Seed Germination.Plants (Basel). 2024 Jul 25;13(15):2051. doi: 10.3390/plants13152051. Plants (Basel). 2024. PMID: 39124169 Free PMC article. Review.
-
First off the mark: early seed germination.J Exp Bot. 2011 Jun;62(10):3289-309. doi: 10.1093/jxb/err030. Epub 2011 Mar 23. J Exp Bot. 2011. PMID: 21430292 Review.
Cited by
-
In-Depth Investigation of Low-Abundance Proteins in Matured and Filling Stages Seeds of Glycine max Employing a Combination of Protamine Sulfate Precipitation and TMT-Based Quantitative Proteomic Analysis.Cells. 2020 Jun 22;9(6):1517. doi: 10.3390/cells9061517. Cells. 2020. PMID: 32580392 Free PMC article.
-
Temperature Regulation of Primary and Secondary Seed Dormancy in Rosa canina L.: Findings from Proteomic Analysis.Int J Mol Sci. 2020 Sep 23;21(19):7008. doi: 10.3390/ijms21197008. Int J Mol Sci. 2020. PMID: 32977616 Free PMC article.
-
Identifying novel fruit-related genes in Arabidopsis thaliana based on the random walk with restart algorithm.PLoS One. 2017 May 4;12(5):e0177017. doi: 10.1371/journal.pone.0177017. eCollection 2017. PLoS One. 2017. PMID: 28472169 Free PMC article.
-
Mitochondrial Proteome Studies in Seeds during Germination.Proteomes. 2016 Jun 21;4(2):19. doi: 10.3390/proteomes4020019. Proteomes. 2016. PMID: 28248229 Free PMC article. Review.
-
Expect the Unexpected Enrichment of "Hidden Proteome" of Seeds and Tubers by Depletion of Storage Proteins.Front Plant Sci. 2016 Jun 1;7:761. doi: 10.3389/fpls.2016.00761. eCollection 2016. Front Plant Sci. 2016. PMID: 27313590 Free PMC article.
References
-
- Acquadro A, Falvo S, Mila S, Giuliano Albo A, Comino C, Moglia A, Lanteri S (2009) Proteomics in globe artichoke: protein extraction and sample complexity reduction by PEG fractionation. Electrophoresis 30: 1594–1602 - PubMed
-
- Ahsan N, Lee DG, Lee SH, Kang KY, Bahk JD, Choi MS, Lee IJ, Renaut J, Lee BH (2007) A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiol Plant 131: 555–570 - PubMed
-
- Allen PS, Benech-Arnold RL, Batlla D, Bradford KJ (2007) Modeling of seed dormancy. InBradford KJ, Nonogaki H, eds, Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 72–112
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

