Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals
- PMID: 25736890
- PMCID: PMC4358017
- DOI: 10.1128/mBio.00037-15
Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals
Abstract
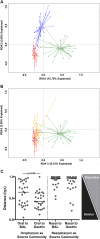
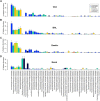
No studies have examined the relationships between bacterial communities along sites of the upper aerodigestive tract of an individual subject. Our objective was to perform an intrasubject and intersite analysis to determine the contributions of two upper mucosal sites (mouth and nose) as source communities for the bacterial microbiome of lower sites (lungs and stomach). Oral wash, bronchoalveolar lavage (BAL) fluid, nasal swab, and gastric aspirate samples were collected from 28 healthy subjects. Extensive analysis of controls and serial intrasubject BAL fluid samples demonstrated that sampling of the lungs by bronchoscopy was not confounded by oral microbiome contamination. By quantitative PCR, the oral cavity and stomach contained the highest bacterial signal levels and the nasal cavity and lungs contained much lower levels. Pyrosequencing of 16S rRNA gene amplicon libraries generated from these samples showed that the oral and gastric compartments had the greatest species richness, which was significantly greater in both than the richness measured in the lungs and nasal cavity. The bacterial communities of the lungs were significantly different from those of the mouth, nose, and stomach, while the greatest similarity was between the oral and gastric communities. However, the bacterial communities of healthy lungs shared significant membership with the mouth, but not the nose, and marked subject-subject variation was noted. In summary, microbial immigration from the oral cavity appears to be the significant source of the lung microbiome during health, but unlike the stomach, the lungs exhibit evidence of selective elimination of Prevotella bacteria derived from the upper airways.
Importance: We have demonstrated that the bacterial communities of the healthy lung overlapped those found in the mouth but were found at lower concentrations, with lower membership and a different community composition. The nasal microbiome, which was distinct from the oral microbiome, appeared to contribute little to the composition of the lung microbiome in healthy subjects. Our studies of the nasal, oral, lung, and stomach microbiomes within an individual illustrate the microbiological continuity of the aerodigestive tract in healthy adults and provide culture-independent microbiological support for the concept that microaspiration is common in healthy individuals.
Copyright © 2015 Bassis et al.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
