Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1
- PMID: 25737329
- PMCID: PMC4418657
- DOI: 10.1002/cbic.201402708
Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1
Abstract
Site-specific incorporation of non-standard amino acids (NSAAs) into proteins opens the way to novel biological insights and applications in biotechnology. Here, we describe the development of a high yielding cell-free protein synthesis (CFPS) platform for NSAA incorporation from crude extracts of genomically recoded Escherichia coli lacking release factor 1. We used genome engineering to construct synthetic organisms that, upon cell lysis, lead to improved extract performance. We targeted five potential negative effectors to be disabled: the nuclease genes rna, rnb, csdA, mazF, and endA. Using our most productive extract from strain MCJ.559 (csdA(-) endA(-)), we synthesized 550±40 μg mL(-1) of modified superfolder green fluorescent protein containing p-acetyl-L-phenylalanine. This yield was increased to ∼1300 μg mL(-1) when using a semicontinuous method. Our work has implications for using whole genome editing for CFPS strain development, expanding the chemistry of biological systems, and cell-free synthetic biology.
Keywords: cell-free protein synthesis; genome engineering; non-standard amino acids; release factor 1; synthetic biology.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
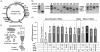
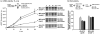
 ), 60 (
), 60 (
 ), and 180 min (■)) of Spinach aptamer plasmid DNA with cell extract, CFPS reagents were added and incubated at 30°C. Maximum mRNA synthesis levels from the mRNA synthesis time course (Figure S2) were compared. At least three independent reactions for each sample were performed, and one standard deviation is shown.
), and 180 min (■)) of Spinach aptamer plasmid DNA with cell extract, CFPS reagents were added and incubated at 30°C. Maximum mRNA synthesis levels from the mRNA synthesis time course (Figure S2) were compared. At least three independent reactions for each sample were performed, and one standard deviation is shown.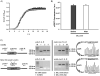

 ), soluble (
), soluble (
 ), and active (■) protein yields of sfGFP and CAT with and without single pAcF. At least three independent CFPS reactions for each sample were performed for 20 h at 30°C for (A) and (C), and one standard deviation is shown.
), and active (■) protein yields of sfGFP and CAT with and without single pAcF. At least three independent CFPS reactions for each sample were performed for 20 h at 30°C for (A) and (C), and one standard deviation is shown.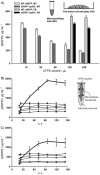
 ), CFPS reagents from the substrate reservoir passively diffuse into the CFPS reaction (inward arrows) through the microdialysis membrane, while by-products are removed from the CFPS reaction (outward arrows). Batches were of 15 (●), 30 (
), CFPS reagents from the substrate reservoir passively diffuse into the CFPS reaction (inward arrows) through the microdialysis membrane, while by-products are removed from the CFPS reaction (outward arrows). Batches were of 15 (●), 30 (
 ), 60 (
), 60 (
 ), 120 (⋄), and 240 μL (▲). At least three independent reactions for each sample were performed at 30°C, and one standard deviation is shown.
), 120 (⋄), and 240 μL (▲). At least three independent reactions for each sample were performed at 30°C, and one standard deviation is shown.References
-
- Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G. Angew Chem Int Ed. 2012;51:2243–2246. - PubMed
- Angew Chem. 2012;124:2286–2289.
- Hong SH, Ntai I, Haimovich AD, Kelleher NL, Isaacs FJ, Jewett MC. ACS Synth Biol. 2014;3:398–409. - PMC - PubMed
- Albayrak C, Swartz JR. Nucleic Acids Res. 2013;41:5949–5963. - PMC - PubMed
- Hayashi Y, Morimoto J, Suga H. ACS Chem Biol. 2012;7:607–613. - PubMed
-
- Zimmerman ES, Heibeck TH, Gill A, Li X, Murray CJ, Madlansacay MR, Tran C, Uter NT, Yin G, Rivers PJ, Yam AY, Wang WD, Steiner AR, Bajad SU, Penta K, Yang W, Hallam TJ, Thanos CD, Sato AK. Bioconjugate Chem. 2014;25:351–361. - PubMed
- Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, Lu Y, Tran H, Seller AJ, Biroc SL, Szydlik A, Pinkstaff JK, Tian F, Sinha SC, Felding-Habermann B, Smider VV, Schultz PG. Proc Natl Acad Sci USA. 2012;109:16101–16106. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

