Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing
- PMID: 25737808
- PMCID: PMC4338798
- DOI: 10.7717/peerj.739
Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing
Abstract
The ability of Shiga toxin-producing Escherichia coli (STEC) to cause severe illness in humans is determined by multiple host factors and bacterial characteristics, including Shiga toxin (Stx) subtype. Given the link between Stx2a subtype and disease severity, we sought to identify the stx subtypes present in whole genome sequences (WGS) of 444 isolates of STEC O157. Difficulties in assembling the stx genes in some strains were overcome by using two complementary bioinformatics methods: mapping and de novo assembly. We compared the WGS analysis with the results obtained using a PCR approach and investigated the diversity within and between the subtypes. All strains of STEC O157 in this study had stx1a, stx2a or stx2c or a combination of these three genes. There was over 99% (442/444) concordance between PCR and WGS. When common source strains were excluded, 236/349 strains of STEC O157 had multiple copies of different Stx subtypes and 54 had multiple copies of the same Stx subtype. Of those strains harbouring multiple copies of the same Stx subtype, 33 had variants between the alleles while 21 had identical copies. Strains harbouring Stx2a only were most commonly found to have multiple alleles of the same subtype (42%). Both the PCR and WGS approach to stx subtyping provided a good level of sensitivity and specificity. In addition, the WGS data also showed there were a significant proportion of strains harbouring multiple alleles of the same Stx subtype associated with clinical disease in England.
Keywords: E. coli; Genomics; O157; Sequencing; Stx.
Conflict of interest statement
Philip M. Ashton, Neil Perry, Kathie A. Grant, Claire Jenkins, and Tim J. Dallman are employees of Public Health England, and Richard Ellis and Liljana Petrovska are employees of Animal Health and Veterinary Laboratories Agency.
Figures

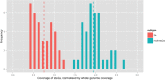
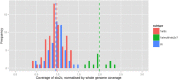



References
-
- Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathogens. 2009;5(5):e739. doi: 10.1371/journal.ppat.1000408. - DOI - PMC - PubMed
-
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

