The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism
- PMID: 25738457
- PMCID: PMC4757431
- DOI: 10.1016/j.cmet.2015.02.008
The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism
Abstract
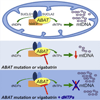
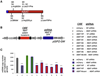
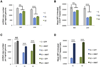
ABAT is a key enzyme responsible for catabolism of principal inhibitory neurotransmitter γ-aminobutyric acid (GABA). We report an essential role for ABAT in a seemingly unrelated pathway, mitochondrial nucleoside salvage, and demonstrate that mutations in this enzyme cause an autosomal recessive neurometabolic disorder and mtDNA depletion syndrome (MDS). We describe a family with encephalomyopathic MDS caused by a homozygous missense mutation in ABAT that results in elevated GABA in subjects' brains as well as decreased mtDNA levels in subjects' fibroblasts. Nucleoside rescue and co-IP experiments pinpoint that ABAT functions in the mitochondrial nucleoside salvage pathway to facilitate conversion of dNDPs to dNTPs. Pharmacological inhibition of ABAT through the irreversible inhibitor Vigabatrin caused depletion of mtDNA in photoreceptor cells that was prevented through addition of dNTPs in cell culture media. This work reveals ABAT as a connection between GABA metabolism and nucleoside metabolism and defines a neurometabolic disorder that includes MDS.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




References
-
- Al-Hussaini A, Faqeih E, El-Hattab AW, Alfadhel M, Asery A, Alsaleem B, Bakhsh E, Ali A, Alasmari A, Lone K, Nahari A, Eyaid W, Al Balwi M, Craig K, Butterworth A, He L, Taylor RW. Clinical and molecular characteristics of mitochondrial DNA depletion syndrome associated with neonatal cholestasis and liver failure. J Pediatr. 2014;164:553–559. e551–e552. - PubMed
-
- Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, Saleh MA, Eyaid W, Hadeel A, He L, Smith F, Yau S, Simcox EM, Miwa S, Donti T, Abu-Amero KK, Wong LJ, Craigen WJ, Graham BH, Scott KL, McFarland R, Taylor RW. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–481. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA125123/CA/NCI NIH HHS/United States
- G0800674/MRC_/Medical Research Council/United Kingdom
- R01 GM098387/GM/NIGMS NIH HHS/United States
- 096919/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- U54 HD083092/HD/NICHD NIH HHS/United States
- 096919/WT_/Wellcome Trust/United Kingdom
- P30 AI036211/AI/NIAID NIH HHS/United States
- S10 RR024574/RR/NCRR NIH HHS/United States
- R01 NS083726/NS/NINDS NIH HHS/United States
- RR024574/RR/NCRR NIH HHS/United States
- AI036211/AI/NIAID NIH HHS/United States
- G0601943/MRC_/Medical Research Council/United Kingdom
- P30 CA125123/CA/NCI NIH HHS/United States
- R01NS083726/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

