Circulating microRNA-196a as a candidate diagnostic biomarker for chronic hepatitis C
- PMID: 25738504
- PMCID: PMC4438874
- DOI: 10.3892/mmr.2015.3386
Circulating microRNA-196a as a candidate diagnostic biomarker for chronic hepatitis C
Abstract
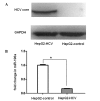
Previous studies have demonstrated the inhibitory effect of microRNA (miR)-196a on hepatitis C virus (HCV) expression in human hepatocytes. However, the clinical implications of aberrant miR-196a expression and the application of circulating miR-196a in the diagnosis and management of chronic hepatitis C (CHC) require further investigation. The present study aimed to examine the possibility of using serum miR-196a as a biomarker for CHC. The Affymetrix miRNA array platform was used for miRNA expression profiling in adenovirus (Ad)-HCV core-infected (HepG2-HCV) and Ad-enhanced green fluorescence protein (EGFP)-infected HepG2 cells (HepG2-control). miR-196a downregulation and levels were analyzed using stem-loop reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis of the sera of 43 patients with CHC and 22 healthy controls. A total of six miRNAs were identified as significantly different (≥ 1.5 fold; P ≤ 0.05) between the two groups. Of note, significant miR-196a downregulation was observed in HepG2-HCV as compared with HepG2‑EGFP. Furthermore, as compared with that of the healthy control group, serum miR-196a was demonstrated to be significantly lower in patients with CHC. In addition, analysis of the receiver operating characteristic (ROC) curve for serum miR-196a revealed an area under the ROC curve of 0.849 (95% confidence interval, 0.756-0.941; P<0.001) with 81.8% sensitivity and 76.7% specificity in discriminating chronic HCV infection from healthy controls at a cut-off value of 6.115 x 10(-5), demonstrating significant diagnostic value for CHC. However, no correlation was identified between serum miR-196a and alanine aminotransferase, aspartate aminotransferase or HCV-RNA. In conclusion, the present study identified circulating miR-196a as a specific and noninvasive candidate biomarker for the diagnosis of CHC.
Figures



Similar articles
-
Dysregulated Serum MicroRNA Expression Profile and Potential Biomarkers in Hepatitis C Virus-infected Patients.Int J Med Sci. 2015 Jul 16;12(7):590-8. doi: 10.7150/ijms.11525. eCollection 2015. Int J Med Sci. 2015. PMID: 26283876 Free PMC article.
-
Circulating microRNAs as predictive biomarkers for liver disease progression of chronic hepatitis C (genotype-4) Egyptian patients.J Med Virol. 2019 Jan;91(1):93-101. doi: 10.1002/jmv.25294. Epub 2018 Sep 24. J Med Virol. 2019. PMID: 30133717
-
Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection.Tumour Biol. 2016 Sep;37(9):12273-12286. doi: 10.1007/s13277-016-5097-8. Epub 2016 Jun 6. Tumour Biol. 2016. PMID: 27271989
-
The Role of MicroRNA in Pathogenesis and as Markers of HCV Chronic Infection.Curr Drug Targets. 2017;18(7):756-765. doi: 10.2174/1389450117666160401125213. Curr Drug Targets. 2017. PMID: 27033188 Review.
-
miR-196a in the carcinogenesis and other disorders with an especial focus on its biomarker capacity.Pathol Res Pract. 2024 Aug;260:155433. doi: 10.1016/j.prp.2024.155433. Epub 2024 Jun 30. Pathol Res Pract. 2024. PMID: 38959626 Review.
Cited by
-
Biomarkers in Detection of Hepatitis C Virus Infection.Pathogens. 2024 Apr 17;13(4):331. doi: 10.3390/pathogens13040331. Pathogens. 2024. PMID: 38668286 Free PMC article. Review.
-
Circulating microRNAs as predictors of response to sofosbuvir + daclatasvir + ribavirin in in HCV genotype-4 Egyptian patients.BMC Gastroenterol. 2022 Dec 3;22(1):499. doi: 10.1186/s12876-022-02485-6. BMC Gastroenterol. 2022. PMID: 36463118 Free PMC article.
-
miRNAs regulate immune response and signaling during hepatitis C virus infection.Eur J Med Res. 2018 Apr 18;23(1):19. doi: 10.1186/s40001-018-0317-x. Eur J Med Res. 2018. PMID: 29669594 Free PMC article. Review.
-
Dysregulated Serum MicroRNA Expression Profile and Potential Biomarkers in Hepatitis C Virus-infected Patients.Int J Med Sci. 2015 Jul 16;12(7):590-8. doi: 10.7150/ijms.11525. eCollection 2015. Int J Med Sci. 2015. PMID: 26283876 Free PMC article.
-
Biological Differentiation of Dampness-Heat Syndromes in Chronic Hepatitis B: From Comparative MicroRNA Microarray Profiling to Biomarker Identification.Evid Based Complement Alternat Med. 2020 Jan 20;2020:7234893. doi: 10.1155/2020/7234893. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 32051688 Free PMC article.
References
-
- Sarrazin C, Wedemeyer H, Cloherty G, et al. Importance of very early HCV RNA kinetics for prediction of treatment outcome of highly effective all oral direct acting antiviral combination therapy. J Virol Methods. 2014;214C:29–32. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

