What controls the hybridization thermodynamics of spherical nucleic acids?
- PMID: 25738968
- PMCID: PMC5490082
- DOI: 10.1021/jacs.5b00670
What controls the hybridization thermodynamics of spherical nucleic acids?
Abstract
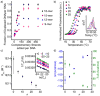
The hybridization of free oligonucleotides to densely packed, oriented arrays of DNA modifying the surfaces of spherical nucleic acid (SNA)-gold nanoparticle conjugates occurs with negative cooperativity; i.e., each binding event destabilizes subsequent binding events. DNA hybridization is thus an ever-changing function of the number of strands already hybridized to the particle. Thermodynamic quantification of this behavior reveals a 3 orders of magnitude decrease in the binding constant for the capture of a free oligonucleotide by an SNA conjugate as the fraction of pre-hybridized strands increases from 0 to ∼30%. Increasing the number of pre-hybridized strands imparts an increasing enthalpic penalty to hybridization that makes binding more difficult, while simultaneously decreasing the entropic penalty to hybridization, which makes binding more favorable. Hybridization of free DNA to an SNA is thus governed by both an electrostatic barrier as the SNA accumulates charge with additional binding events and an effect consistent with allostery, where hybridization at certain sites on an SNA modify the binding affinity at a distal site through conformational changes to the remaining single strands. Leveraging these insights allows for the design of conjugates that hybridize free strands with significantly higher efficiencies, some of which approach 100%.
Figures



References
-
- Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. - PubMed
-
- Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477. - PMC - PubMed
- Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, Mirkin CA. Anal Chem. 2012;84:2062. - PMC - PubMed
- Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, Malin D, Strekalova E, Cryns VL, Cheng C, Mirkin CA, Thaxton CS. Proc Natl Acad Sci U S A. 2014;111:17104. - PMC - PubMed
- Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. ACS Nano. 2009;3:2147. - PMC - PubMed
-
- Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027. - PubMed
- Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, Daniel WL, Scott AW, Rotz MW, Meade TJ, Giljohann DA, Mirkin CA, Stegh AH. Sci Transl Med. 2013;5:209–152. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

