A rapid molecular approach for chromosomal phasing
- PMID: 25739099
- PMCID: PMC4349636
- DOI: 10.1371/journal.pone.0118270
A rapid molecular approach for chromosomal phasing
Abstract
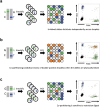
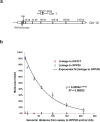
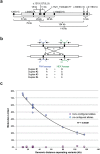
Determining the chromosomal phase of pairs of sequence variants - the arrangement of specific alleles as haplotypes - is a routine challenge in molecular genetics. Here we describe Drop-Phase, a molecular method for quickly ascertaining the phase of pairs of DNA sequence variants (separated by 1-200 kb) without cloning or manual single-molecule dilution. In each Drop-Phase reaction, genomic DNA segments are isolated in tens of thousands of nanoliter-sized droplets together with allele-specific fluorescence probes, in a single reaction well. Physically linked alleles partition into the same droplets, revealing their chromosomal phase in the co-distribution of fluorophores across droplets. We demonstrated the accuracy of this method by phasing members of trios (revealing 100% concordance with inheritance information), and demonstrate a common clinical application by phasing CFTR alleles at genomic distances of 11-116 kb in the genomes of cystic fibrosis patients. Drop-Phase is rapid (requiring less than 4 hours), scalable (to hundreds of samples), and effective at long genomic distances (200 kb).
Conflict of interest statement
Figures

References
-
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nat Genet 2002; 32: 432–437. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

