Regulated eukaryotic DNA replication origin firing with purified proteins
- PMID: 25739503
- PMCID: PMC4874468
- DOI: 10.1038/nature14285
Regulated eukaryotic DNA replication origin firing with purified proteins
Abstract
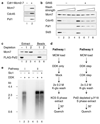
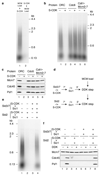
Eukaryotic cells initiate DNA replication from multiple origins, which must be tightly regulated to promote precise genome duplication in every cell cycle. To accomplish this, initiation is partitioned into two temporally discrete steps: a double hexameric minichromosome maintenance (MCM) complex is first loaded at replication origins during G1 phase, and then converted to the active CMG (Cdc45-MCM-GINS) helicase during S phase. Here we describe the reconstitution of budding yeast DNA replication initiation with 16 purified replication factors, made from 42 polypeptides. Origin-dependent initiation recapitulates regulation seen in vivo. Cyclin-dependent kinase (CDK) inhibits MCM loading by phosphorylating the origin recognition complex (ORC) and promotes CMG formation by phosphorylating Sld2 and Sld3. Dbf4-dependent kinase (DDK) promotes replication by phosphorylating MCM, and can act either before or after CDK. These experiments define the minimum complement of proteins, protein kinase substrates and co-factors required for regulated eukaryotic DNA replication.
Figures



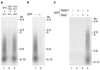
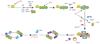






Comment in
-
Molecular biology: DNA replication reconstructed.Nature. 2015 Mar 26;519(7544):418-9. doi: 10.1038/519418a. Nature. 2015. PMID: 25810199 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

