Liver-directed lentiviral gene therapy in a dog model of hemophilia B
- PMID: 25739762
- PMCID: PMC5669486
- DOI: 10.1126/scitranslmed.aaa1405
Liver-directed lentiviral gene therapy in a dog model of hemophilia B
Abstract
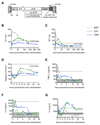
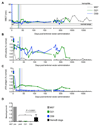
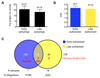
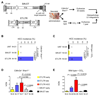
We investigated the efficacy of liver-directed gene therapy using lentiviral vectors in a large animal model of hemophilia B and evaluated the risk of insertional mutagenesis in tumor-prone mouse models. We showed that gene therapy using lentiviral vectors targeting the expression of a canine factor IX transgene in hepatocytes was well tolerated and provided a stable long-term production of coagulation factor IX in dogs with hemophilia B. By exploiting three different mouse models designed to amplify the consequences of insertional mutagenesis, we showed that no genotoxicity was detected with these lentiviral vectors. Our findings suggest that lentiviral vectors may be an attractive candidate for gene therapy targeted to the liver and may be potentially useful for the treatment of hemophilia.
Copyright © 2015, American Association for the Advancement of Science.
Conflict of interest statement
Figures
References
-
- Bolton-Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361:1801–1809. - PubMed
-
- High KH, Nathwani A, Spencer T, Lillicrap D. Current status of haemophilia gene therapy. Haemophilia. 2014;20(Suppl 4):43–49. - PubMed
-
- Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379:1447–1456. - PubMed
-
- Astermark J, Santagostino E, Keith Hoots W. Clinical issues in inhibitors. Haemophilia. 2010;16(Suppl 5):54–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

