New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource
- PMID: 25740547
- PMCID: PMC4350103
- DOI: 10.1038/srep08771
New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource
Abstract
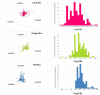
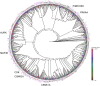
Using whole-cell phenotypic assays, the GlaxoSmithKline high-throughput screening (HTS) diversity set of 1.8 million compounds was screened against the three kinetoplastids most relevant to human disease, i.e. Leishmania donovani, Trypanosoma cruzi and Trypanosoma brucei. Secondary confirmatory and orthogonal intracellular anti-parasiticidal assays were conducted, and the potential for non-specific cytotoxicity determined. Hit compounds were chemically clustered and triaged for desirable physicochemical properties. The hypothetical biological target space covered by these diversity sets was investigated through bioinformatics methodologies. Consequently, three anti-kinetoplastid chemical boxes of ~200 compounds each were assembled. Functional analyses of these compounds suggest a wide array of potential modes of action against kinetoplastid kinases, proteases and cytochromes as well as potential host-pathogen targets. This is the first published parallel high throughput screening of a pharma compound collection against kinetoplastids. The compound sets are provided as an open resource for future lead discovery programs, and to address important research questions.
Figures


Comment in
-
Parasite infection: Parasite-targeting compound sets identified and ready to go.Nat Rev Drug Discov. 2015 Apr;14(4):238. doi: 10.1038/nrd4588. Nat Rev Drug Discov. 2015. PMID: 25829282 No abstract available.
References
-
- Pedrique B. et al. The drug and vaccine landscape for neglected diseases (2000–2011): a systematic assessment. Lancet Glob. Health 1, e371–e379 (2013). - PubMed
-
- World Health Organization. Working to overcome the global impact of neglected tropical diseases – first WHO report on neglected tropical diseases. http://whqlibdoc.who.int/publications/2010/9789241564090_eng.pdf (2010) (Date of access: 20th November 2014).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

