Alterations in hemagglutinin receptor-binding specificity accompany the emergence of highly pathogenic avian influenza viruses
- PMID: 25741006
- PMCID: PMC4442535
- DOI: 10.1128/JVI.03304-14
Alterations in hemagglutinin receptor-binding specificity accompany the emergence of highly pathogenic avian influenza viruses
Abstract
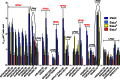
Highly pathogenic avian influenza viruses (HPAIVs) of hemagglutinin H5 and H7 subtypes emerge after introduction of low-pathogenic avian influenza viruses (LPAIVs) from wild birds into poultry flocks, followed by subsequent circulation and evolution. The acquisition of multiple basic amino acids at the endoproteolytical cleavage site of the hemagglutinin (HA) is a molecular indicator for high pathogenicity, at least for infections of gallinaceous poultry. Apart from the well-studied significance of the multibasic HA cleavage site, there is only limited knowledge on other alterations in the HA and neuraminidase (NA) molecules associated with changes in tropism during the emergence of HPAIVs from LPAIVs. We hypothesized that changes in tropism may require alterations of the sialyloligosaccharide specificities of HA and NA. To test this hypothesis, we compared a number of LPAIVs and HPAIVs for their HA-mediated binding and NA-mediated desialylation of a set of synthetic receptor analogs, namely, α2-3-sialylated oligosaccharides. NA substrate specificity correlated with structural groups of NAs and did not correlate with pathogenic potential of the virus. In contrast, all HPAIVs differed from LPAIVs by a higher HA receptor-binding affinity toward the trisaccharides Neu5Acα2-3Galβ1-4GlcNAcβ (3'SLN) and Neu5Acα2-3Galβ1-3GlcNAcβ (SiaLe(c)) and by the ability to discriminate between the nonfucosylated and fucosylated sialyloligosaccharides 3'SLN and Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ (SiaLe(x)), respectively. These results suggest that alteration of the receptor-binding specificity accompanies emergence of the HPAIVs from their low-pathogenic precursors.
Importance: Here, we have found for the first time correlations of receptor-binding properties of the HA with a highly pathogenic phenotype of poultry viruses. Our study suggests that enhanced receptor-binding affinity of HPAIVs for a typical "poultry-like" receptor, 3'SLN, is provided by substitutions in the receptor-binding site of HA which appeared in HA of LPAIVs in the course of transmission of LPAIVs from wild waterfowl into poultry flocks, with subsequent adaptation in poultry. The identification of LPAIVs with receptor characteristics of HPAIVs argues that the sialic acid-binding specificity of the HA may be used as a novel phenotypic marker of HPAIVs.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Emergence and Adaptation of a Novel Highly Pathogenic H7N9 Influenza Virus in Birds and Humans from a 2013 Human-Infecting Low-Pathogenic Ancestor.J Virol. 2018 Jan 2;92(2):e00921-17. doi: 10.1128/JVI.00921-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29070694 Free PMC article.
-
Structure and receptor binding specificity of hemagglutinin H13 from avian influenza A virus H13N6.J Virol. 2013 Aug;87(16):9077-85. doi: 10.1128/JVI.00235-13. Epub 2013 Jun 12. J Virol. 2013. PMID: 23760233 Free PMC article.
-
Genetic and antigenic characterization of H5 and H7 avian influenza viruses isolated from migratory waterfowl in Mongolia from 2017 to 2019.Virus Genes. 2020 Aug;56(4):472-479. doi: 10.1007/s11262-020-01764-2. Epub 2020 May 19. Virus Genes. 2020. PMID: 32430568 Free PMC article.
-
Influenza pathobiology and pathogenesis in avian species.Curr Top Microbiol Immunol. 2014;385:221-42. doi: 10.1007/82_2014_385. Curr Top Microbiol Immunol. 2014. PMID: 25015786 Review.
-
Hemagglutinin Subtype Specificity and Mechanisms of Highly Pathogenic Avian Influenza Virus Genesis.Viruses. 2022 Jul 19;14(7):1566. doi: 10.3390/v14071566. Viruses. 2022. PMID: 35891546 Free PMC article. Review.
Cited by
-
Complex N-glycans are important for interspecies transmission of H7 influenza A viruses.J Virol. 2024 Apr 16;98(4):e0194123. doi: 10.1128/jvi.01941-23. Epub 2024 Mar 12. J Virol. 2024. PMID: 38470143 Free PMC article.
-
Host Receptors of Influenza Viruses and Coronaviruses-Molecular Mechanisms of Recognition.Vaccines (Basel). 2020 Oct 6;8(4):587. doi: 10.3390/vaccines8040587. Vaccines (Basel). 2020. PMID: 33036202 Free PMC article. Review.
-
Amino acid residues at positions 222 and 227 of the hemagglutinin together with the neuraminidase determine binding of H5 avian influenza viruses to sialyl Lewis X.Arch Virol. 2016 Feb;161(2):307-16. doi: 10.1007/s00705-015-2660-3. Epub 2015 Nov 5. Arch Virol. 2016. PMID: 26542967 Free PMC article.
-
Avian Influenza H5N6 Viruses Exhibit Differing Pathogenicities and Transmissibilities in Mammals.Sci Rep. 2017 Nov 24;7(1):16280. doi: 10.1038/s41598-017-16139-1. Sci Rep. 2017. PMID: 29176564 Free PMC article.
-
Genetic Characterization and Pathogenesis of Avian Influenza Virus H7N3 Isolated from Spot-Billed Ducks in South Korea, Early 2019.Viruses. 2021 May 7;13(5):856. doi: 10.3390/v13050856. Viruses. 2021. PMID: 34067187 Free PMC article.
References
-
- Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus ADME. 2005. Characterization of a novel influenza a virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi:10.1128/JVI.79.5.2814-2822.2005. - DOI - PMC - PubMed
-
- World Organisation for Animal Health (OIE). 2005. Avian influenza, chapter 2.7.12. In OIE manual of diagnostic tests and vaccines for terrestrial animals, 5th ed World Organisation for Animal Health (OIE), Paris, France.
-
- World Organisation for Animal Health (OIE). 2007. Avian influenza, chapter 2.7.12. In Terrestrial animal health code, 16th ed World Organisation for Animal Health, Paris, France: http://www.oie.int/doc/ged/d6430.pdf.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

