Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis
- PMID: 25741013
- PMCID: PMC4403485
- DOI: 10.1128/JVI.03652-14
Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis
Abstract
Ebola virus VP35 inhibits alpha/beta interferon production and functions as a viral polymerase cofactor. Previously, the 8-kDa cytoplasmic dynein light chain (LC8) was demonstrated to interact with VP35, but the functional consequences were unclear. Here we demonstrate that the interaction is direct and of high affinity and that binding stabilizes the VP35 N-terminal oligomerization domain and enhances viral RNA synthesis. Mutational analysis demonstrates that VP35 interaction is required for the functional effects of LC8.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

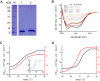
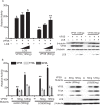
Similar articles
-
Virus and host interactions critical for filoviral RNA synthesis as therapeutic targets.Antiviral Res. 2019 Feb;162:90-100. doi: 10.1016/j.antiviral.2018.12.006. Epub 2018 Dec 11. Antiviral Res. 2019. PMID: 30550800 Free PMC article. Review.
-
Crystal structure of human LC8 bound to a peptide from Ebola virus VP35.J Microbiol. 2021 Apr;59(4):410-416. doi: 10.1007/s12275-021-0641-7. Epub 2021 Feb 25. J Microbiol. 2021. PMID: 33630249
-
The Host E3-Ubiquitin Ligase TRIM6 Ubiquitinates the Ebola Virus VP35 Protein and Promotes Virus Replication.J Virol. 2017 Aug 24;91(18):e00833-17. doi: 10.1128/JVI.00833-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679761 Free PMC article.
-
Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function.J Virol. 2010 Oct;84(20):10581-91. doi: 10.1128/JVI.00925-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686031 Free PMC article.
-
DRBP76 associates with Ebola virus VP35 and suppresses viral polymerase function.J Infect Dis. 2011 Nov;204 Suppl 3(Suppl 3):S911-8. doi: 10.1093/infdis/jir343. J Infect Dis. 2011. PMID: 21987769 Free PMC article.
Cited by
-
Virus and host interactions critical for filoviral RNA synthesis as therapeutic targets.Antiviral Res. 2019 Feb;162:90-100. doi: 10.1016/j.antiviral.2018.12.006. Epub 2018 Dec 11. Antiviral Res. 2019. PMID: 30550800 Free PMC article. Review.
-
Current status of small molecule drug development for Ebola virus and other filoviruses.Curr Opin Virol. 2019 Apr;35:42-56. doi: 10.1016/j.coviro.2019.03.001. Epub 2019 Apr 16. Curr Opin Virol. 2019. PMID: 31003196 Free PMC article. Review.
-
Crystal Structure of the Marburg Virus VP35 Oligomerization Domain.J Virol. 2017 Jan 3;91(2):e01085-16. doi: 10.1128/JVI.01085-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847355 Free PMC article.
-
Dynein Light Chain LC8 Is Required for RNA Polymerase I-Mediated Transcription in Trypanosoma brucei, Facilitating Assembly and Promoter Binding of Class I Transcription Factor A.Mol Cell Biol. 2015 Oct 12;36(1):95-107. doi: 10.1128/MCB.00705-15. Print 2016 Jan 1. Mol Cell Biol. 2015. PMID: 26459761 Free PMC article.
-
Single-Cell Profiling of Ebola Virus Disease In Vivo Reveals Viral and Host Dynamics.Cell. 2020 Nov 25;183(5):1383-1401.e19. doi: 10.1016/j.cell.2020.10.002. Epub 2020 Nov 6. Cell. 2020. PMID: 33159858 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI081914/AI/NIAID NIH HHS/United States
- T32-CA09547-37/CA/NCI NIH HHS/United States
- R01 AI107056/AI/NIAID NIH HHS/United States
- U19 AI070489/AI/NIAID NIH HHS/United States
- R01AI081914/AI/NIAID NIH HHS/United States
- T32 AI007163/AI/NIAID NIH HHS/United States
- R01AI059536/AI/NIAID NIH HHS/United States
- R01 AI059536/AI/NIAID NIH HHS/United States
- U19 AI109945/AI/NIAID NIH HHS/United States
- P01 AI120943/AI/NIAID NIH HHS/United States
- U19AI109945/AI/NIAID NIH HHS/United States
- R01AI107056/AI/NIAID NIH HHS/United States
- U19 AI109664/AI/NIAID NIH HHS/United States
- U19AI109664/AI/NIAID NIH HHS/United States
- T32 CA009547/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

