Summarizing and exploring data of a decade of cytokinin-related transcriptomics
- PMID: 25741346
- PMCID: PMC4330702
- DOI: 10.3389/fpls.2015.00029
Summarizing and exploring data of a decade of cytokinin-related transcriptomics
Abstract
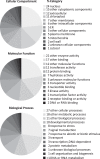
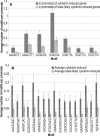
The genome-wide transcriptional response of the model organism Arabidopsis thaliana to cytokinin has been investigated by different research groups as soon as large-scale transcriptomic techniques became affordable. Over the last 10 years many transcriptomic datasets related to cytokinin have been generated using different technological platforms, some of which are published only in databases, culminating in an RNA sequencing experiment. Two approaches have been made to establish a core set of cytokinin-regulated transcripts by meta-analysis of these datasets using different preferences regarding their selection. Here we add another meta-analysis derived from an independent microarray platform (CATMA), combine all the meta-analyses available with RNAseq data in order to establish an advanced core set of cytokinin-regulated transcripts, and compare the results with the regulation of orthologous rice genes by cytokinin. We discuss the functions of some of the less known cytokinin-regulated genes indicating areas deserving further research to explore cytokinin function. Finally, we investigate the promoters of the core set of cytokinin-induced genes for the abundance and distribution of known cytokinin-responsive cis elements and identify a set of novel candidate motifs.
Keywords: cis element; cytokinin; gene regulation; hormone action; meta-analysis; regulatory network; signal transduction; transcriptomics.
Figures


Similar articles
-
Gene regulation by cytokinin in Arabidopsis.Front Plant Sci. 2012 Jan 31;3:8. doi: 10.3389/fpls.2012.00008. eCollection 2012. Front Plant Sci. 2012. PMID: 22639635 Free PMC article.
-
In planta analysis of a cis-regulatory cytokinin response motif in Arabidopsis and identification of a novel enhancer sequence.Plant Cell Physiol. 2013 Jul;54(7):1079-92. doi: 10.1093/pcp/pct060. Epub 2013 Apr 24. Plant Cell Physiol. 2013. PMID: 23620480
-
Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades.Plant J. 2005 Oct;44(2):314-33. doi: 10.1111/j.1365-313X.2005.02530.x. Plant J. 2005. PMID: 16212609
-
Cytokinin Signaling Downstream of the His-Asp Phosphorelay Network: Cytokinin-Regulated Genes and Their Functions.Front Plant Sci. 2020 Nov 17;11:604489. doi: 10.3389/fpls.2020.604489. eCollection 2020. Front Plant Sci. 2020. PMID: 33329676 Free PMC article. Review.
-
Role of the Cytokinin-Activated Type-B Response Regulators in Hormone Crosstalk.Plants (Basel). 2020 Jan 30;9(2):166. doi: 10.3390/plants9020166. Plants (Basel). 2020. PMID: 32019090 Free PMC article. Review.
Cited by
-
ARR17 controls dioecy in Populus by repressing B-class MADS-box gene expression.Philos Trans R Soc Lond B Biol Sci. 2022 May 9;377(1850):20210217. doi: 10.1098/rstb.2021.0217. Epub 2022 Mar 21. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35306887 Free PMC article.
-
Cytokinin Determines Thiol-Mediated Arsenic Tolerance and Accumulation.Plant Physiol. 2016 Jun;171(2):1418-26. doi: 10.1104/pp.16.00372. Epub 2016 Apr 18. Plant Physiol. 2016. PMID: 27208271 Free PMC article.
-
Cytokinin-Regulated Expression of Arabidopsis thaliana PAP Genes and Its Implication for the Expression of Chloroplast-Encoded Genes.Biomolecules. 2020 Dec 11;10(12):1658. doi: 10.3390/biom10121658. Biomolecules. 2020. PMID: 33322466 Free PMC article.
-
Signal Integration in Plant Abiotic Stress Responses via Multistep Phosphorelay Signaling.Front Plant Sci. 2021 Feb 17;12:644823. doi: 10.3389/fpls.2021.644823. eCollection 2021. Front Plant Sci. 2021. PMID: 33679861 Free PMC article. Review.
-
Prunus Knotted-like Genes: Genome-Wide Analysis, Transcriptional Response to Cytokinin in Micropropagation, and Rootstock Transformation.Int J Mol Sci. 2023 Feb 3;24(3):3046. doi: 10.3390/ijms24033046. Int J Mol Sci. 2023. PMID: 36769369 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

