Conformational dynamics and antigenicity in the disordered malaria antigen merozoite surface protein 2
- PMID: 25742002
- PMCID: PMC4351039
- DOI: 10.1371/journal.pone.0119899
Conformational dynamics and antigenicity in the disordered malaria antigen merozoite surface protein 2
Abstract
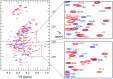
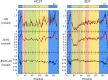
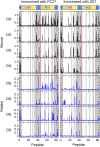
Merozoite surface protein 2 (MSP2) of Plasmodium falciparum is an abundant, intrinsically disordered protein that is GPI-anchored to the surface of the invasive blood stage of the malaria parasite. Recombinant MSP2 has been trialled as a component of a malaria vaccine, and is one of several disordered proteins that are candidates for inclusion in vaccines for malaria and other diseases. Nonetheless, little is known about the implications of protein disorder for the development of an effective antibody response. We have therefore undertaken a detailed analysis of the conformational dynamics of the two allelic forms of MSP2 (3D7 and FC27) using NMR spectroscopy. Chemical shifts and NMR relaxation data indicate that conformational and dynamic properties of the N- and C-terminal conserved regions in the two forms of MSP2 are essentially identical, but significant variation exists between and within the central variable regions. We observe a strong relationship between the conformational dynamics and the antigenicity of MSP2, as assessed with antisera to recombinant MSP2. Regions of increased conformational order in MSP2, including those in the conserved regions, are more strongly antigenic, while the most flexible regions are minimally antigenic. This suggests that modifications that increase conformational order may offer a means to tune the antigenicity of MSP2 and other disordered antigens, with implications for vaccine design.
Conflict of interest statement
Figures

References
-
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. 2005; J Mol Recognit 18: 343–384. - PubMed
-
- Feng ZP, Zhang X, Han P, Arora N, Anders RF, Norton RS. Abundance of intrinsically unstructured proteins in P. falciparum and other apicomplexan parasite proteomes. 2006; Mol Biochem Parasitol 150: 256–267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

