Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients
- PMID: 25742471
- PMCID: PMC4402458
- DOI: 10.1038/bjc.2015.80
Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients
Abstract
Introduction: Recent discoveries in cancer research have revealed a plethora of clinically actionable mutations that provide therapeutic, prognostic and predictive benefit to patients. The feasibility of screening mutations as part of the routine clinical care of patients remains relatively unexplored as the demonstration of massively parallel sequencing (MPS) of tumours in the general population is required to assess its value towards the health-care system.
Methods: Cancer 2015 study is a large-scale, prospective, multisite cohort of newly diagnosed cancer patients from Victoria, Australia with 1094 patients recruited. MPS was performed using the Illumina TruSeq Amplicon Cancer Panel.
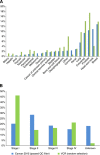
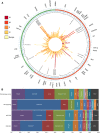
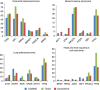
Results: Overall, 854 patients were successfully sequenced for 48 common cancer genes. Accurate determination of clinically relevant mutations was possible including in less characterised cancer types; however, technical limitations including formalin-induced sequencing artefacts were uncovered. Applying strict filtering criteria, clinically relevant mutations were identified in 63% of patients, with 26% of patients displaying a mutation with therapeutic implications. A subset of patients was validated for canonical mutations using the Agena Bioscience MassARRAY system with 100% concordance. Whereas the prevalence of mutations was consistent with other institutionally based series for some tumour streams (breast carcinoma and colorectal adenocarcinoma), others were different (lung adenocarcinoma and head and neck squamous cell carcinoma), which has significant implications for health economic modelling of particular targeted agents. Actionable mutations in tumours not usually thought to harbour such genetic changes were also identified.
Conclusions: Reliable delivery of a diagnostic assay able to screen for a range of actionable mutations in this cohort was achieved, opening unexpected avenues for investigation and treatment of cancer patients.
Figures
References
-
- Bourgon R, Lu S, Yan Y, Lackner MR, Wang W, Weigman V, Wang D, Guan Y, Ryner L, Koeppen H, Patel R, Hampton GM, Amler LC, Wang Y. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next generation sequencing. Clin Cancer Res. 2014;20 (8:2080–2091. - PubMed
-
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2 (5:401–404. - PMC - PubMed
-
- Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61 (1:64–71. - PubMed
-
- Do H, Wong SQ, Li J, Dobrovic A. Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem. 2013;59 (9:1376–1383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

