Quantitative studies of mRNA recruitment to the eukaryotic ribosome
- PMID: 25742741
- PMCID: PMC4458453
- DOI: 10.1016/j.biochi.2015.02.017
Quantitative studies of mRNA recruitment to the eukaryotic ribosome
Abstract
The process of peptide bond synthesis by ribosomes is conserved between species, but the initiation step differs greatly between the three kingdoms of life. This is illustrated by the evolution of roughly an order of magnitude more initiation factor mass found in humans compared with bacteria. Eukaryotic initiation of translation is comprised of a number of sub-steps: (i) recruitment of an mRNA and initiator methionyl-tRNA to the 40S ribosomal subunit; (ii) migration of the 40S subunit along the 5' UTR to locate the initiation codon; and (iii) recruitment of the 60S subunit to form the 80S initiation complex. Although the mechanism and regulation of initiation has been studied for decades, many aspects of the pathway remain unclear. In this review, I will focus discussion on what is known about the mechanism of mRNA selection and its recruitment to the 40S subunit. I will summarize how the 43S preinitiation complex (PIC) is formed and stabilized by interactions between its components. I will discuss what is known about the mechanism of mRNA selection by the eukaryotic initiation factor 4F (eIF4F) complex and how the selected mRNA is recruited to the 43S PIC. The regulation of this process by secondary structure located in the 5' UTR of an mRNA will also be discussed. Finally, I present a possible kinetic model with which to explain the process of mRNA selection and recruitment to the eukaryotic ribosome.
Keywords: 43S PIC; eIF4F; mRNA recruitment.
Copyright © 2015 Elsevier B.V. and Société française de biochimie et biologie Moléculaire (SFBBM). All rights reserved.
Figures
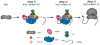




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

