The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression
- PMID: 25742952
- PMCID: PMC4350857
- DOI: 10.1186/s12943-015-0318-0
The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression
Abstract
Background: Long noncoding RNAs (lncRNAs) have emerged recently as a new class of genes that regulate cellular processes, such as cell growth and apoptosis. The SPRY4 intronic transcript 1 (SPRY4-IT1) is a 708-bp lncRNA on chromosome 5 with a potential functional role in tumorigenesis. The clinical significance of SPRY4-IT1 and the effect of SPRY4-IT1 on cancer progression are unclear.
Methods: Quantitative reverse transcriptase PCR (qRT-PCR) was performed to investigate the expression of SPRY4-IT1 in 48 breast cancer tissues and four breast cancer cell lines. Gain and loss of function approaches were used to investigate the biological role of SPRY4-IT1 in vitro. Microarray bioinformatics analysis was performed to identify the putative targets of SPRY4-IT1, which were further verified by rescue experiments, and by western blotting and qRT-PCR.
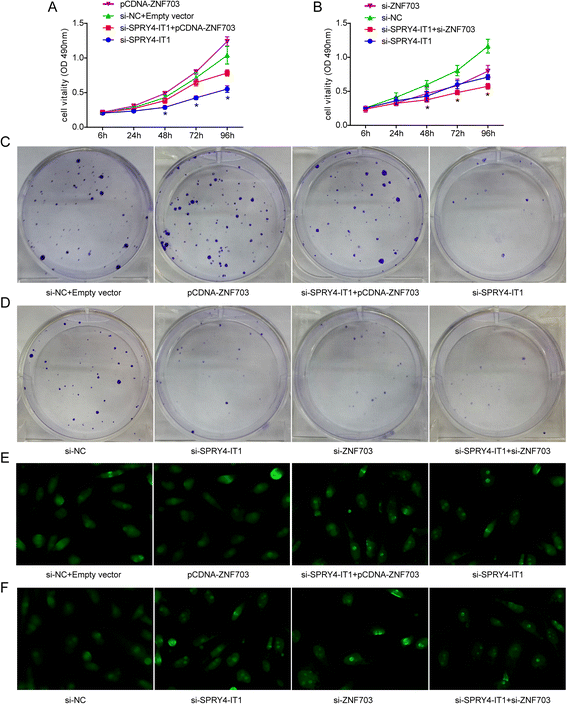
Results: SPRY4-IT1 expression was significantly upregulated in 48 breast cancer tumor tissues comparedwith normal tissues. Additionally, increased SPRY4-IT1 expression was found to be associated with a larger tumor size and an advanced pathological stage in breast cancer patients. The knockdown of SPRY4-IT1 significantly suppressed proliferation and caused apoptosis of breast cancer cells in vitro. Furthermore, we discovered that ZNF703 was a target of SPRY4-IT1 and was downregulated by SPRY4-IT1 knockdown. Moreover, we provide the first demonstration that ZNF703 plays an oncogenic role in ER (-) breast carcinoma cells.
Conclusions: SPRY4-IT1 is a novel prognostic biomarker and a potential therapeutic candidate for breast cancer.
Figures





References
-
- Bosma AJ, Weigelt B, Lambrechts AC, Verhagen OJ, Pruntel R, Hart AA, et al. Detection of circulating breast tumor cells by differential expression of marker genes. Clin Cancer Res. 2002;8:1871–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

