Neighboring Genes Show Correlated Evolution in Gene Expression
- PMID: 25743543
- PMCID: PMC4476153
- DOI: 10.1093/molbev/msv053
Neighboring Genes Show Correlated Evolution in Gene Expression
Abstract
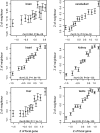
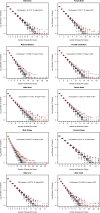
When considering the evolution of a gene's expression profile, we commonly assume that this is unaffected by its genomic neighborhood. This is, however, in contrast to what we know about the lack of autonomy between neighboring genes in gene expression profiles in extant taxa. Indeed, in all eukaryotic genomes genes of similar expression-profile tend to cluster, reflecting chromatin level dynamics. Does it follow that if a gene increases expression in a particular lineage then the genomic neighbors will also increase in their expression or is gene expression evolution autonomous? To address this here we consider evolution of human gene expression since the human-chimp common ancestor, allowing for both variation in estimation of current expression level and error in Bayesian estimation of the ancestral state. We find that in all tissues and both sexes, the change in gene expression of a focal gene on average predicts the change in gene expression of neighbors. The effect is highly pronounced in the immediate vicinity (<100 kb) but extends much further. Sex-specific expression change is also genomically clustered. As genes increasing their expression in humans tend to avoid nuclear lamina domains and be enriched for the gene activator 5-hydroxymethylcytosine, we conclude that, most probably owing to chromatin level control of gene expression, a change in gene expression of one gene likely affects the expression evolution of neighbors, what we term expression piggybacking, an analog of hitchhiking.
Keywords: gene clustering; gene expression evolution; sex-biased evolution.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Barton RA, Venditti C. Rapid evolution of the cerebellum in humans and other great apes. Curr Biol. 2014;24:2440–2444. - PubMed
-
- Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2007;23:480–484. - PubMed
-
- Birnbaum K, Shasha D, Wang J, Jung J, Lambert G, Galbraith D, Benfey P. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. - PubMed
-
- Blumenthal T, Evans D, Link C, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu W, Duke K, Kiraly M, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

