Structure-function analysis of USP1: insights into the role of Ser313 phosphorylation site and the effect of cancer-associated mutations on autocleavage
- PMID: 25744535
- PMCID: PMC4326527
- DOI: 10.1186/s12943-015-0311-7
Structure-function analysis of USP1: insights into the role of Ser313 phosphorylation site and the effect of cancer-associated mutations on autocleavage
Abstract
Background: In complex with its cofactor UAF1, the USP1 deubiquitinase plays an important role in cellular processes related to cancer, including the response to DNA damage. The USP1/UAF1 complex is emerging as a novel target in cancer therapy, but several aspects of its function and regulation remain to be further clarified. These include the role of the serine 313 phosphorylation site, the relative contribution of different USP1 sequence motifs to UAF1 binding, and the potential effect of cancer-associated mutations on USP1 regulation by autocleavage.
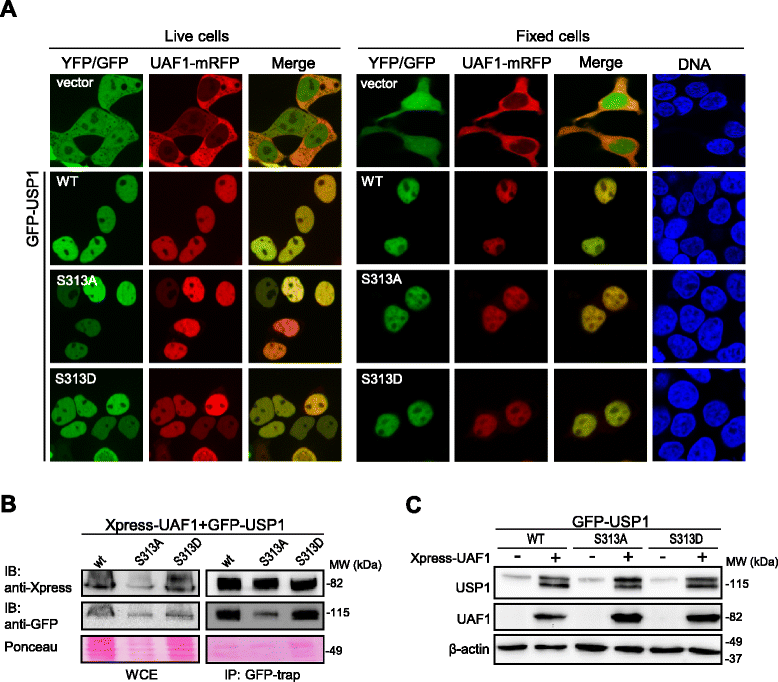
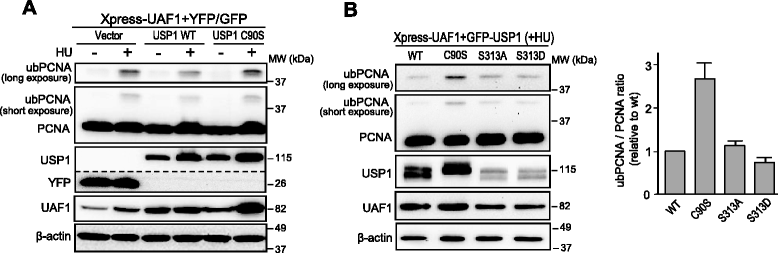
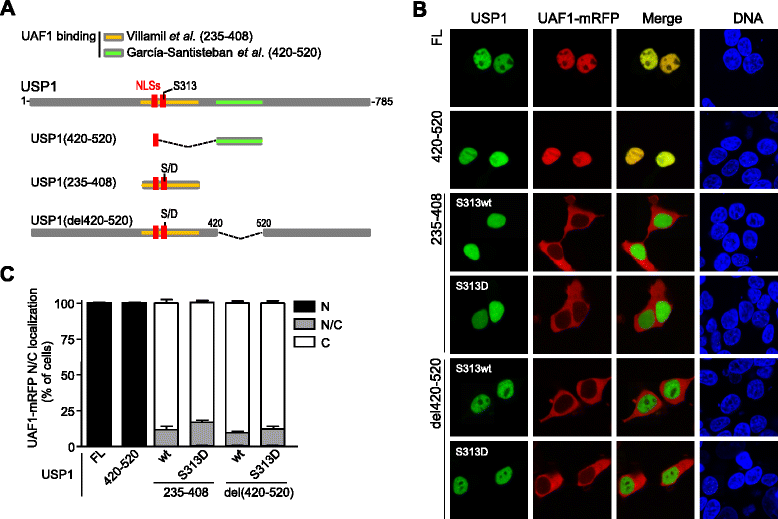
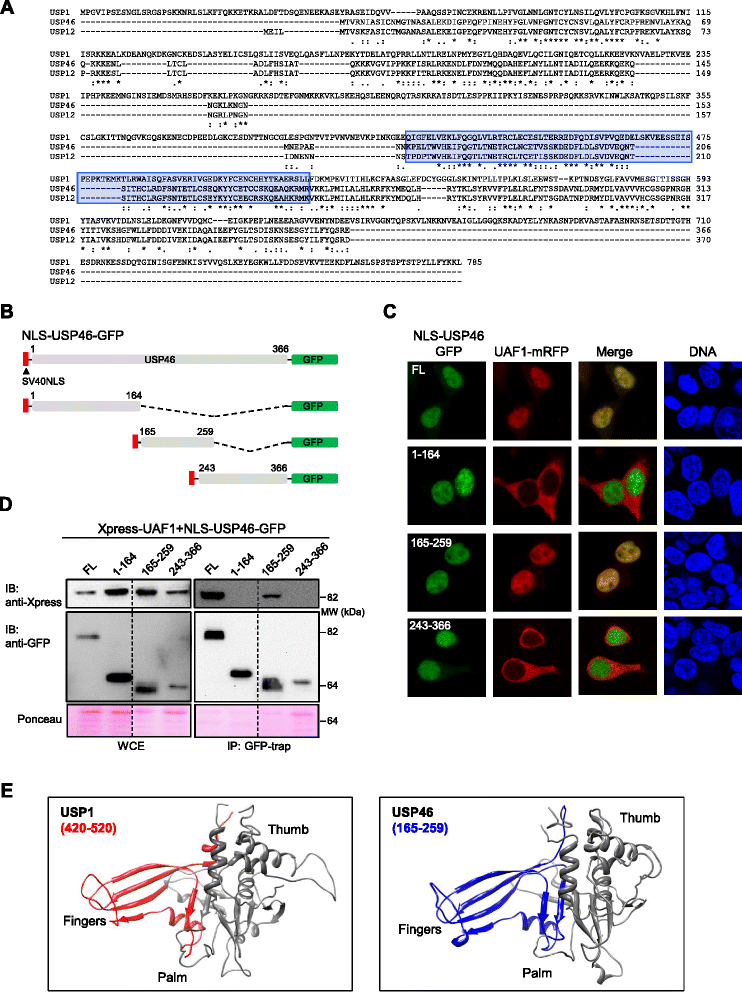
Methods: We have generated a large set of USP1 structural variants, including a catalytically inactive form (C90S), non-phosphorylatable (S313A) and phosphomimetic (S313D) mutants, deletion mutants lacking potential UAF1 binding sites, a mutant (GG/AA) unable to undergo autocleavage at the well-characterized G670/G671 diglycine motif, and four USP1 mutants identified in tumor samples that cluster around this cleavage site (G667A, L669P, K673T and A676T). Using cell-based assays, we have determined the ability of these mutants to bind UAF1, to reverse DNA damage-induced monoubiquitination of PCNA, and to undergo autocleavage.
Results: A non-phosphorylatable S313A mutant of USP1 retained the ability to bind UAF1 and to reverse PCNA ubiquitination in cell-based assays. Regardless of the presence of a phosphomimetic S313D mutation, deletion of USP1 fragment 420-520 disrupted UAF1 binding, as determined using a nuclear relocation assay. The UAF1 binding site in a second UAF1-interacting DUB, USP46, was mapped to a region homologous to USP1(420-520). Regarding USP1 autocleavage, co-expression of the C90S and GG/AA mutants did not result in cleavage, while the cancer-associated mutation L669P was found to reduce cleavage efficiency.
Conclusions: USP1 phosphorylation at S313 is not critical for PCNA deubiquitination, neither for binding to UAF1 in a cellular environment. In this context, USP1 amino acid motif 420-520 is necessary and sufficient for UAF1 binding. This motif, and a homologous amino acid segment that mediates USP46 binding to UAF1, map to the Fingers sub-domain of these DUBs. On the other hand, our results support the view that USP1 autocleavage may occur in cis, and can be altered by a cancer-associated mutation.
Figures

Similar articles
-
Mutations in the 'Fingers' subdomain of the deubiquitinase USP1 modulate its function and activity.FEBS J. 2016 Mar;283(5):929-46. doi: 10.1111/febs.13648. Epub 2016 Feb 3. FEBS J. 2016. PMID: 26758085
-
Serine phosphorylation is critical for the activation of ubiquitin-specific protease 1 and its interaction with WD40-repeat protein UAF1.Biochemistry. 2012 Nov 13;51(45):9112-23. doi: 10.1021/bi300845s. Epub 2012 Nov 1. Biochemistry. 2012. PMID: 23116119 Free PMC article.
-
Two nuclear localization signals in USP1 mediate nuclear import of the USP1/UAF1 complex.PLoS One. 2012;7(6):e38570. doi: 10.1371/journal.pone.0038570. Epub 2012 Jun 6. PLoS One. 2012. PMID: 22701671 Free PMC article.
-
The WD40-repeat protein-containing deubiquitinase complex: catalysis, regulation, and potential for therapeutic intervention.Cell Biochem Biophys. 2013 Sep;67(1):111-26. doi: 10.1007/s12013-013-9637-1. Cell Biochem Biophys. 2013. PMID: 23797609 Free PMC article. Review.
-
Ubiquitin-specific peptidase 1: assessing its role in cancer therapy.Clin Exp Med. 2023 Nov;23(7):2953-2966. doi: 10.1007/s10238-023-01075-4. Epub 2023 Apr 24. Clin Exp Med. 2023. PMID: 37093451 Review.
Cited by
-
Regulation of Deubiquitinating Enzymes by Post-Translational Modifications.Int J Mol Sci. 2020 Jun 4;21(11):4028. doi: 10.3390/ijms21114028. Int J Mol Sci. 2020. PMID: 32512887 Free PMC article. Review.
-
A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.J Struct Biol. 2016 Dec;196(3):437-447. doi: 10.1016/j.jsb.2016.09.011. Epub 2016 Sep 17. J Struct Biol. 2016. PMID: 27650958 Free PMC article.
-
Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy.Int J Mol Sci. 2020 Apr 6;21(7):2548. doi: 10.3390/ijms21072548. Int J Mol Sci. 2020. PMID: 32268558 Free PMC article. Review.
-
Branched-chain amino acids catabolism and cancer progression: focus on therapeutic interventions.Front Oncol. 2023 Aug 10;13:1220638. doi: 10.3389/fonc.2023.1220638. eCollection 2023. Front Oncol. 2023. PMID: 37637065 Free PMC article. Review.
-
Characterization of the deubiquitination activity and substrate specificity of the chicken ubiquitin-specific protease 1/USP associated factor 1 complex.PLoS One. 2017 Nov 1;12(11):e0186535. doi: 10.1371/journal.pone.0186535. eCollection 2017. PLoS One. 2017. PMID: 29091922 Free PMC article.
References
-
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

