Lysosomal protein turnover contributes to the acquisition of TGFβ-1 induced invasive properties of mammary cancer cells
- PMID: 25744631
- PMCID: PMC4339013
- DOI: 10.1186/s12943-015-0313-5
Lysosomal protein turnover contributes to the acquisition of TGFβ-1 induced invasive properties of mammary cancer cells
Abstract
Background: Normal epithelial cells and carcinoma cells can acquire invasiveness by epithelial-to-mesenchymal transition (EMT), a process of considerable cellular remodeling. The endosomal/lysosomal compartment is a principal site of intracellular protein degradation. Lysosomal cathepsin proteases are secreted during cancer progression. The established pro-metastatic role of specific cysteine cathepsins has until now been ascribed to their contribution to extracellular matrix remodeling. We hypothesized that cysteine cathepsins affect transforming growth factor β-1 (TGFβ-1)-induced EMT of normal and malignant mammary epithelial cells.
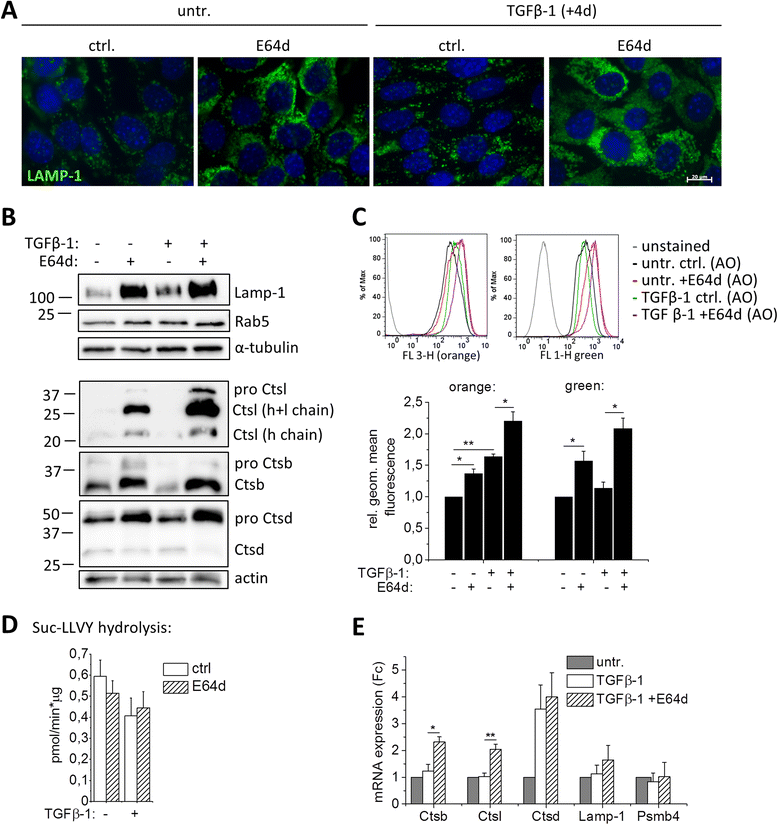
Methods: The role of lysosomal proteolysis in TGFβ-1-induced EMT and invasion was investigated in a normal and a novel malignant murine mammary epithelial cell line. The contribution of cysteine cathepsins was determined by addition of the general cysteine cathepsin inhibitor E64d. Hallmarks of EMT were analyzed by molecular- and cell-biologic analyses including real-time cell migration/invasion assays. A quantitative proteome comparison using stable isotopic labeling with amino acids in culture (SILAC) showed the effect of E64d on TGFβ-1 induced proteome changes. Lysosomal patterning and junctional adhesion molecule A (Jam-a) localization and abundance were analyzed by immunofluorescence.
Results: We found increased lysosome activity during EMT of malignant mammary epithelial cells. Cysteine cathepsin inhibition had no effect on the induction of the TGFβ-1-induced EMT program on transcriptional level. Protease inhibition did not affect invasion of TGFβ-1 treated normal mammary epithelial cells, but reduced the invasion of murine breast cancer cells. Remarkably, reduced invasion was also evident if E64d was removed 24 h before the invasion assay in order to allow for recovery of cathepsin activity. Proteome analyses revealed a high abundance of lysosomal enzymes and lysosome-associated proteins in cancer cells treated with TGFβ-1 and E64d. An accumulation of those proteins and of lysosomal vesicles was further confirmed by independent methods. Interestingly, E64d caused lysosomal accumulation of Jam-a, a tight junction component facilitating epithelial cell-cell adhesion.
Conclusion: Our results demonstrate an important role of lysosomal proteolysis in cellular remodeling during EMT and a pivotal contribution of lysosomal cysteine cathepsins to TGFβ-1 induced acquisition of breast cancer cell invasiveness. These findings provide an additional rationale to use cathepsin inhibitors to stall tumor metastasis.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

