Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV
- PMID: 25746232
- PMCID: PMC4845099
- DOI: 10.1021/cb500917m
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV
Abstract
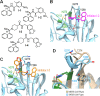
The Middle East Respiratory Syndrome coronavirus (MERS-CoV) papain-like protease (PLpro) blocking loop 2 (BL2) structure differs significantly from that of SARS-CoV PLpro, where it has been proven to play a crucial role in SARS-CoV PLpro inhibitor binding. Four SARS-CoV PLpro lead inhibitors were tested against MERS-CoV PLpro, none of which were effective against MERS-CoV PLpro. Structure and sequence alignments revealed that two residues, Y269 and Q270, responsible for inhibitor binding to SARS-CoV PLpro, were replaced by T274 and A275 in MERS-CoV PLpro, making critical binding interactions difficult to form for similar types of inhibitors. High-throughput screening (HTS) of 25 000 compounds against both PLpro enzymes identified a small fragment-like noncovalent dual inhibitor. Mode of inhibition studies by enzyme kinetics and competition surface plasmon resonance (SPR) analyses suggested that this compound acts as a competitive inhibitor with an IC50 of 6 μM against MERS-CoV PLpro, indicating that it binds to the active site, whereas it acts as an allosteric inhibitor against SARS-CoV PLpro with an IC50 of 11 μM. These results raised the possibility that inhibitor recognition specificity of MERS-CoV PLpro may differ from that of SARS-CoV PLpro. In addition, inhibitory activity of this compound was selective for SARS-CoV and MERS-CoV PLpro enzymes over two human homologues, the ubiquitin C-terminal hydrolases 1 and 3 (hUCH-L1 and hUCH-L3).
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Anderson L. J.; Baric R. S. (2012) Emerging human coronaviruses--disease potential and preparedness. N. Engl. J. Med. 367, 1850–1852. - PubMed
-
- Zaki A. M.; van Boheemen S.; Bestebroer T. M.; Osterhaus A. D.; Fouchier R. A. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. - PubMed
-
- de Groot R. J.; Baker S. C.; Baric R. S.; Brown C. S.; Drosten C.; Enjuanes L.; Fouchier R. A.; Galiano M.; Gorbalenya A. E.; Memish Z. A.; Perlman S.; Poon L. L.; Snijder E. J.; Stephens G. M.; Woo P. C.; Zaki A. M.; Zambon M.; Ziebuhr J. (2013) Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 87, 7790–7792. - PMC - PubMed
-
- World Health Organization (2014). Middle East respiratory syndrome coronavirus (MERS-CoV) – update. http://www.who.int/csr/don/2014_07_23_mers/en/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Miscellaneous

