Structural characterisation of the virulence-associated protein VapG from the horse pathogen Rhodococcus equi
- PMID: 25746683
- PMCID: PMC4518536
- DOI: 10.1016/j.vetmic.2015.01.027
Structural characterisation of the virulence-associated protein VapG from the horse pathogen Rhodococcus equi
Abstract
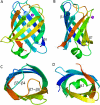
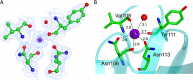
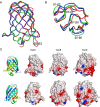
Virulence and host range in Rhodococcus equi depends on the variable pathogenicity island of their virulence plasmids. Notable gene products are a family of small secreted virulence-associated proteins (Vaps) that are critical to intramacrophagic proliferation. Equine-adapted strains, which cause severe pyogranulomatous pneumonia in foals, produce a cell-associated VapA that is necessary for virulence, alongside five other secreted homologues. In the absence of biochemical insight, attention has turned to the structures of these proteins to develop a functional hypothesis. Recent studies have described crystal structures for VapD and a truncate of the VapA orthologue of porcine-adapted strains, VapB. Here, we crystallised the full-length VapG and determined its structure by molecular replacement. Electron density corresponding to the N-terminal domain was not visible suggesting that it is disordered. The protein core adopted a compact elliptical, anti-parallel β-barrel fold with β1-β2-β3-β8-β5-β6-β7-β4 topology decorated by a single peripheral α-helix unique to this family. The high glycine content of the protein allows close packing of secondary structural elements. Topologically, the surface has no indentations that indicate a nexus for molecular interactions. The distribution of polar and apolar groups on the surface of VapG is markedly uneven. One-third of the surface is dominated by exposed apolar side-chains, with no ionisable and only four polar side-chains exposed, giving rise to an expansive flat hydrophobic surface. Other surface regions are more polar, especially on or near the α-helix and a belt around the centre of the β-barrel. Possible functional significance of these recent structures is discussed.
Keywords: Protein Structure; Rhodococcus equi; VapA; VapG; Virulence-associated protein.
Copyright © 2015. Published by Elsevier B.V.
Figures


Similar articles
-
Characterization of virulence plasmid types in Rhodococcus equi isolates from foals, pigs, humans and soil in Hungary.Vet Microbiol. 2002 Sep 24;88(4):377-84. doi: 10.1016/s0378-1135(02)00157-8. Vet Microbiol. 2002. PMID: 12220812
-
Molecular characterization of Rhodococcus equi from horse-breeding farms by means of multiplex PCR for the vap gene family.Curr Microbiol. 2009 Apr;58(4):399-403. doi: 10.1007/s00284-009-9370-6. Epub 2009 Feb 10. Curr Microbiol. 2009. PMID: 19205798
-
Genomic analysis of a novel Rhodococcus (Prescottella) equi isolate from a bovine host.Arch Microbiol. 2019 Nov;201(9):1317-1321. doi: 10.1007/s00203-019-01695-z. Epub 2019 Jul 13. Arch Microbiol. 2019. PMID: 31302711 Free PMC article.
-
Rhodococcus equi: the many facets of a pathogenic actinomycete.Vet Microbiol. 2013 Nov 29;167(1-2):9-33. doi: 10.1016/j.vetmic.2013.06.016. Epub 2013 Jul 5. Vet Microbiol. 2013. PMID: 23993705 Review.
-
Rhodococcus equi: clinical manifestations, virulence, and immunity.J Vet Intern Med. 2011 Nov-Dec;25(6):1221-30. doi: 10.1111/j.1939-1676.2011.00804.x. Epub 2011 Oct 7. J Vet Intern Med. 2011. PMID: 22092609 Review.
Cited by
-
A multiomics approach to identify host-microbe alterations associated with infection severity in diabetic foot infections: a pilot study.NPJ Biofilms Microbiomes. 2021 Mar 22;7(1):29. doi: 10.1038/s41522-021-00202-x. NPJ Biofilms Microbiomes. 2021. PMID: 33753735 Free PMC article.
-
The N-terminal domain is required for cell surface localisation of VapA, a member of the Vap family of Rhodococcus equi virulence proteins.PLoS One. 2024 Feb 29;19(2):e0298900. doi: 10.1371/journal.pone.0298900. eCollection 2024. PLoS One. 2024. PMID: 38421980 Free PMC article.
-
Identification of a VapA virulence factor functional homolog in Rhodococcus equi isolates housing the pVAPB plasmid.PLoS One. 2018 Oct 4;13(10):e0204475. doi: 10.1371/journal.pone.0204475. eCollection 2018. PLoS One. 2018. PMID: 30286098 Free PMC article.
-
Differential Effects of Rhodococcus equi Virulence-Associated Proteins on Macrophages and Artificial Lipid Membranes.Microbiol Spectr. 2023 Feb 14;11(2):e0341722. doi: 10.1128/spectrum.03417-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36786596 Free PMC article.
-
Vaccination of Mice with Virulence-Associated Protein G (VapG) Antigen Confers Partial Protection against Rhodococcus equi Infection through Induced Humoral Immunity.Front Microbiol. 2017 May 11;8:857. doi: 10.3389/fmicb.2017.00857. eCollection 2017. Front Microbiol. 2017. PMID: 28553279 Free PMC article.
References
-
- Asoh N., Watanabe H., Fines-Guyon M., Watanabe K., Oishi K., Kositsakulchai W., Sanchai T., Kunsuikmengrai K., Kahintapong S., Khantawa B., Tharavichitkul P., Sirisanthana T., Nagatake T. Emergence of Rifampin-resistant Rhodococcus equi with several types of mutations in the rhoB gene among AIDS patients in northern Thailand. J. Clin. Microbiol. 2003:2337–2340. - PMC - PubMed
-
- Benoit S., Benachour A., Taouji S., Auffray Y., Hartke A. Induction of vap genes encoded by the virulence plasmid of Rhodococcus equi during acid tolerance response. Res. Microbiol. 2001;152:439–449. - PubMed
-
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

