Regulation of circRNA biogenesis
- PMID: 25746834
- PMCID: PMC4615371
- DOI: 10.1080/15476286.2015.1020271
Regulation of circRNA biogenesis
Abstract
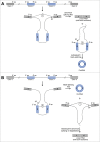
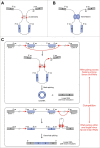
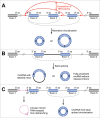
Unlike linear RNAs terminated with 5' caps and 3' tails, circular RNAs are characterized by covalently closed loop structures with neither 5' to 3' polarity nor polyadenylated tail. This intrinsic characteristic has led to the general under-estimation of the existence of circular RNAs in previous polyadenylated transcriptome analyses. With the advent of specific biochemical and computational approaches, a large number of circular RNAs from back-spliced exons (circRNAs) have been identified in various cell lines and across different species. Recent studies have uncovered that back-splicing requires canonical spliceosomal machinery and can be facilitated by both complementary sequences and specific protein factors. In this review, we highlight our current understanding of the regulation of circRNA biogenesis, including both the competition between splicing and back-splicing and the previously under-appreciated alternative circularization.
Keywords: alternative circularization; back-splicing; circular RNA; circularization; complementary sequence; splicing.
Figures

References
-
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73: 3852-6; PMID:1069269; http://dx.doi.org/10.1073/pnas.73.11.3852 - DOI - PMC - PubMed
-
- Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell 1980; 19: 313–9; PMID:6986989; http://dx.doi.org/10.1016/0092-8674(80)90505-X - DOI - PubMed
-
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558–60; PMID:2429192; http://dx.doi.org/10.1038/323558a0 - DOI - PubMed
-
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607–13; PMID:1991322; http://dx.doi.org/10.1016/0092-8674(91)90244-S - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
