The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations
- PMID: 25748176
- PMCID: PMC4352061
- DOI: 10.1371/journal.pone.0119355
The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations
Abstract
Background: High-throughput sequencing has enabled detailed insights into complex microbial environments, including the human gut microbiota. The accuracy of the sequencing data however, is reliant upon appropriate storage of the samples prior to DNA extraction. The aim of this study was to conduct the first MiSeq sequencing investigation into the effects of faecal storage on the microbiota, compared to fresh samples. Culture-based analysis was also completed.
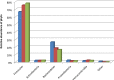
Methods: Seven faecal samples were collected from healthy adults. Samples were separated into fresh (DNA extracted immediately), snap frozen on dry ice and frozen for 7 days at -80°C prior to DNA extraction or samples frozen at -80°C for 7 days before DNA extraction. Sequencing was completed on the Illumina MiSeq platform. Culturing of total aerobes, anaerobes and bifidobacteria was also completed.
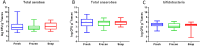
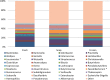
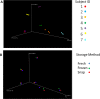
Results: No significant differences at phylum or family levels between the treatment groups occurred. At genus level only Faecalibacterium and Leuconostoc were significantly different in the fresh samples compared to the snap frozen group (p = 0.0298; p = 0.0330 respectively). Diversity analysis indicated that samples clustered based on the individual donor, rather than by storage group. No significant differences occurred in the culture-based analysis between the fresh, snap or -80°C frozen samples.
Conclusions: Using the MiSeq platform coupled with culture-based analysis, this study highlighted that limited significant changes in microbiota occur following rapid freezing of faecal samples prior to DNA extraction. Thus, rapid freezing of samples prior to DNA extraction and culturing, preserves the integrity of the microbiota.
Conflict of interest statement
Figures
References
-
- Blaut M, Collins M, Welling G, Dore J, Van Loo J, De Vos W. Molecular biological methods for studying the gut microbiota: the EU human gut flora project. British Journal of Nutrition. 2002;87(S2):S203– S11. - PubMed
-
- Cani PD. Gut microbiota and obesity: lessons from the microbiome. Briefings in functional genomics. 2013:elt014. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

