Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition
- PMID: 25748651
- PMCID: PMC4359640
- DOI: 10.1016/j.cell.2015.01.050
Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition
Abstract
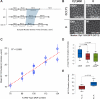
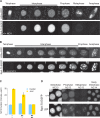
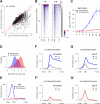
A conserved feature of the midblastula transition (MBT) is a requirement for a functional DNA replication checkpoint to coordinate cell-cycle remodeling and zygotic genome activation (ZGA). We have investigated what triggers this checkpoint during Drosophila embryogenesis. We find that the magnitude of the checkpoint scales with the quantity of transcriptionally engaged DNA. Measuring RNA polymerase II (Pol II) binding at 20 min intervals over the course of ZGA reveals that the checkpoint coincides with widespread de novo recruitment of Pol II that precedes and does not require a functional checkpoint. This recruitment drives slowing or stalling of DNA replication at transcriptionally engaged loci. Reducing Pol II recruitment in zelda mutants both reduces replication stalling and bypasses the requirement for a functional checkpoint. This suggests a model where the checkpoint functions as a feedback mechanism to remodel the cell cycle in response to nascent ZGA.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


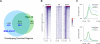

Comment in
-
Transcription gets to the checkpoint.Cell. 2015 Mar 12;160(6):1043-4. doi: 10.1016/j.cell.2015.02.051. Cell. 2015. PMID: 25768900
References
-
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. - PubMed
-
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

