Criteria for selecting microhaplotypes: mixture detection and deconvolution
- PMID: 25750707
- PMCID: PMC4351693
- DOI: 10.1186/s13323-014-0018-3
Criteria for selecting microhaplotypes: mixture detection and deconvolution
Abstract
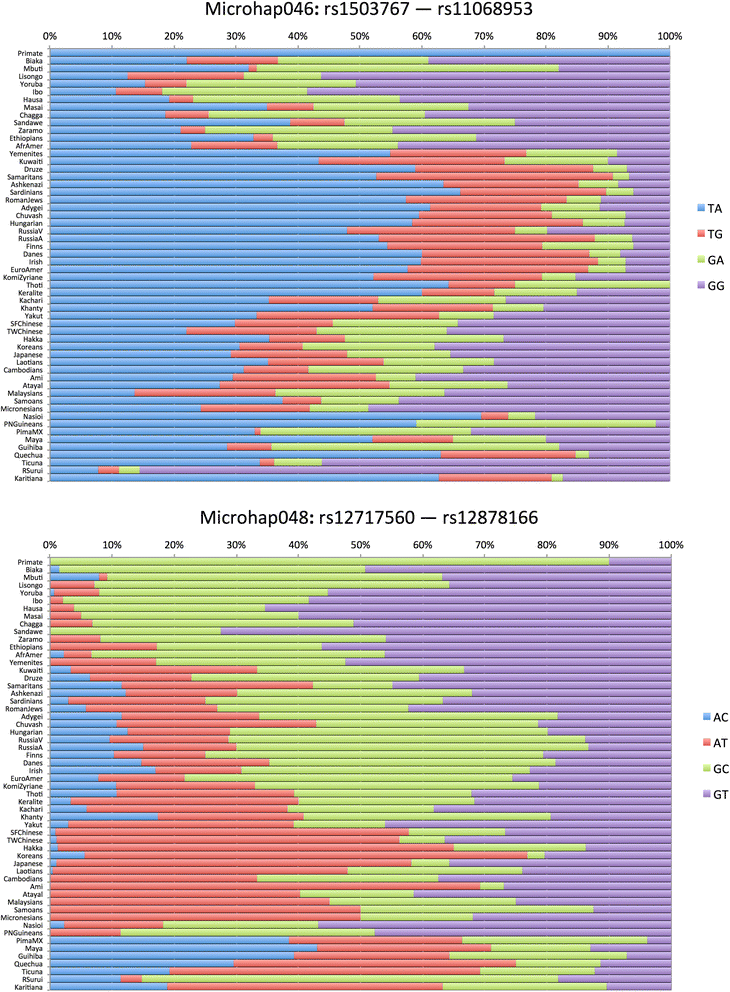
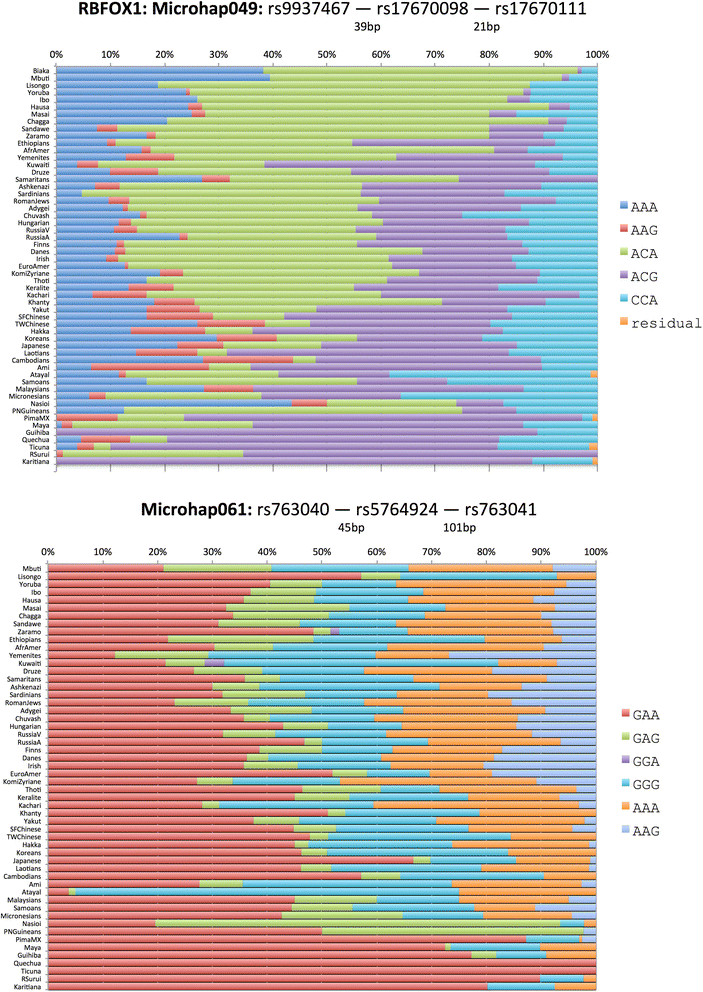
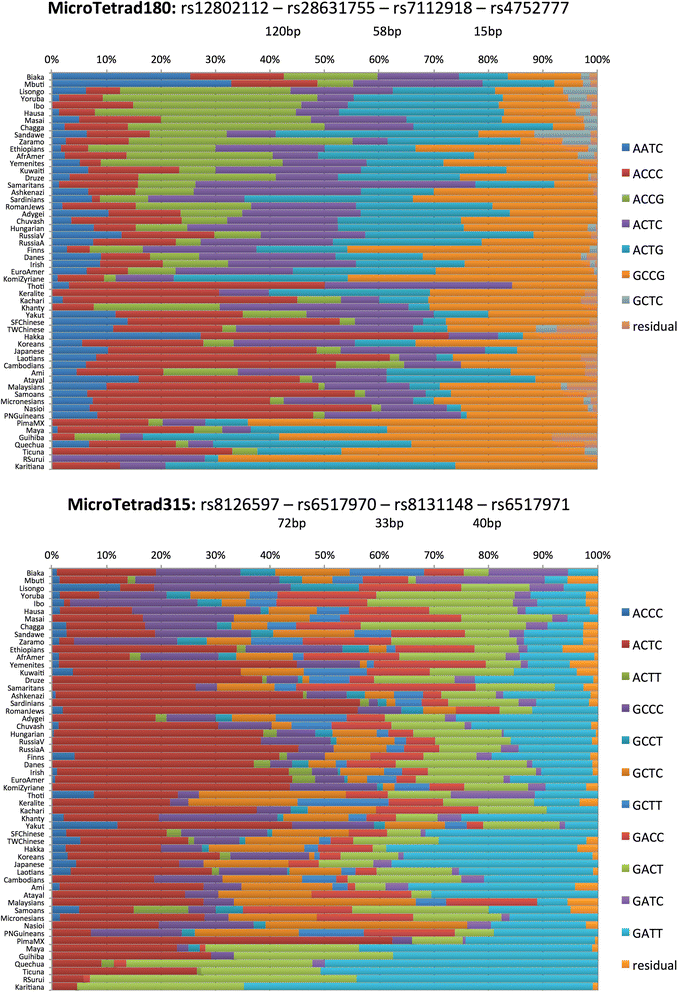
Background: DNA sequencing is likely to become a standard typing method in forensics in the near future. We define a microhaplotype to be a locus with two or more single nucleotide polymorphisms (SNPs) that occur within a short segment of DNA (e.g., 200 bp) that can be covered by a single sequence run and collectively define a multiallelic locus. Microhaplotypes can be highly informative for many forensic questions, including detection of mixtures of two or more sources in a DNA sample, a common problem in forensic practice.
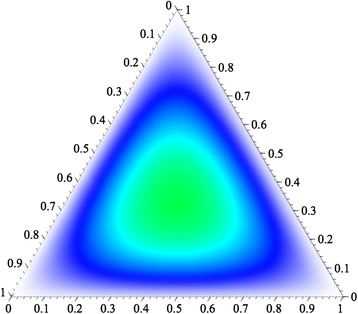
Results: When all alleles are equally frequent, the probability of detecting three or more alleles in a mixture is at maximum. The classical population genetics concept of effective number of alleles at a locus, termed Ae, converts the unequal allele frequencies at a locus into a value that is equivalent to some number of equally frequent alleles, allowing microhaplotype loci to be ranked. The expectations for the ability to qualitatively detect mixtures are given for different integer values of Ae, and the cumulative probabilities of detecting mixtures based on testing multiple microhaps are shown to exceed 95% with as few as five loci with average Ae values of even slightly greater than 3.0.
Conclusions: Microhaplotypes with Ae values of >3 will be exceedingly useful in ordinary forensic practice. Based on our studies, 3-SNP microhaplotypes will sometimes meet this criterion, but 4-SNP microhaplotypes can even exceed this criterion and have values >4.
Keywords: DNA mixtures; Forensic identification; Microhaplotype; Population genetics.
Figures

References
-
- Budowle B, Moretti TR, Niezgoda SJ, Brown BL. Proceedings of the Second European Symposium on Human Identification. Madison, WI: Promega Corporation; 1998. CODIS and PCR-based short tandem repeat loci: Law Enforcement Tools; pp. 73–88.
-
- Federal Bureau of Investigation: FBI–Combined DNA Index System [http://www.fbi.gov/about-us/lab/biometric-analysis/codis]
LinkOut - more resources
Full Text Sources
Other Literature Sources

