A versatile modular vector system for rapid combinatorial mammalian genetics
- PMID: 25751063
- PMCID: PMC4396471
- DOI: 10.1172/JCI79743
A versatile modular vector system for rapid combinatorial mammalian genetics
Abstract
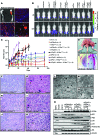
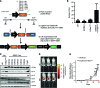
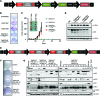
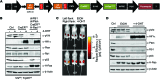
Here, we describe the multiple lentiviral expression (MuLE) system that allows multiple genetic alterations to be introduced simultaneously into mammalian cells. We created a toolbox of MuLE vectors that constitute a flexible, modular system for the rapid engineering of complex polycistronic lentiviruses, allowing combinatorial gene overexpression, gene knockdown, Cre-mediated gene deletion, or CRISPR/Cas9-mediated (where CRISPR indicates clustered regularly interspaced short palindromic repeats) gene mutation, together with expression of fluorescent or enzymatic reporters for cellular assays and animal imaging. Examples of tumor engineering were used to illustrate the speed and versatility of performing combinatorial genetics using the MuLE system. By transducing cultured primary mouse cells with single MuLE lentiviruses, we engineered tumors containing up to 5 different genetic alterations, identified genetic dependencies of molecularly defined tumors, conducted genetic interaction screens, and induced the simultaneous CRISPR/Cas9-mediated knockout of 3 tumor-suppressor genes. Intramuscular injection of MuLE viruses expressing oncogenic H-RasG12V together with combinations of knockdowns of the tumor suppressors cyclin-dependent kinase inhibitor 2A (Cdkn2a), transformation-related protein 53 (Trp53), and phosphatase and tensin homolog (Pten) allowed the generation of 3 murine sarcoma models, demonstrating that genetically defined autochthonous tumors can be rapidly generated and quantitatively monitored via direct injection of polycistronic MuLE lentiviruses into mouse tissues. Together, our results demonstrate that the MuLE system provides genetic power for the systematic investigation of the molecular mechanisms that underlie human diseases.
Figures

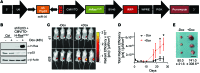


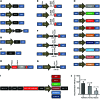
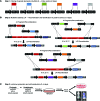
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

