Assembly and specific recognition of k29- and k33-linked polyubiquitin
- PMID: 25752577
- PMCID: PMC4386031
- DOI: 10.1016/j.molcel.2015.01.042
Assembly and specific recognition of k29- and k33-linked polyubiquitin
Abstract
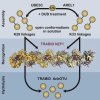
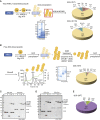
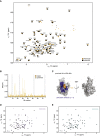
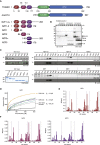
Protein ubiquitination regulates many cellular processes via attachment of structurally and functionally distinct ubiquitin (Ub) chains. Several atypical chain types have remained poorly characterized because the enzymes mediating their assembly and receptors with specific binding properties have been elusive. We found that the human HECT E3 ligases UBE3C and AREL1 assemble K48/K29- and K11/K33-linked Ub chains, respectively, and can be used in combination with DUBs to generate K29- and K33-linked chains for biochemical and structural analyses. Solution studies indicate that both chains adopt open and dynamic conformations. We further show that the N-terminal Npl4-like zinc finger (NZF1) domain of the K29/K33-specific deubiquitinase TRABID specifically binds K29/K33-linked diUb, and a crystal structure of this complex explains TRABID specificity and suggests a model for chain binding by TRABID. Our work uncovers linkage-specific components in the Ub system for atypical K29- and K33-linked Ub chains, providing tools to further understand these unstudied posttranslational modifications.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Make them, break them, and catch them: studying rare ubiquitin chains.Mol Cell. 2015 Apr 2;58(1):1-2. doi: 10.1016/j.molcel.2015.03.024. Mol Cell. 2015. PMID: 25839429
References
-
- Al-Hakim A.K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D.R. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008;411:249–260. - PubMed
-
- Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. - PubMed
-
- Bremm A., Komander D. Emerging roles for Lys11-linked polyubiquitin in cellular regulation. Trends Biochem. Sci. 2011;36:355–363. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

