The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements
- PMID: 25752748
- PMCID: PMC4381529
- DOI: 10.1101/gr.185272.114
The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements
Abstract
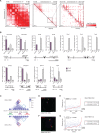
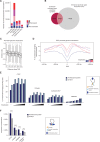
The mammalian genome harbors up to one million regulatory elements often located at great distances from their target genes. Long-range elements control genes through physical contact with promoters and can be recognized by the presence of specific histone modifications and transcription factor binding. Linking regulatory elements to specific promoters genome-wide is currently impeded by the limited resolution of high-throughput chromatin interaction assays. Here we apply a sequence capture approach to enrich Hi-C libraries for >22,000 annotated mouse promoters to identify statistically significant, long-range interactions at restriction fragment resolution, assigning long-range interacting elements to their target genes genome-wide in embryonic stem cells and fetal liver cells. The distal sites contacting active genes are enriched in active histone modifications and transcription factor occupancy, whereas inactive genes contact distal sites with repressive histone marks, demonstrating the regulatory potential of the distal elements identified. Furthermore, we find that coregulated genes cluster nonrandomly in spatial interaction networks correlated with their biological function and expression level. Interestingly, we find the strongest gene clustering in ES cells between transcription factor genes that control key developmental processes in embryogenesis. The results provide the first genome-wide catalog linking gene promoters to their long-range interacting elements and highlight the complex spatial regulatory circuitry controlling mammalian gene expression.
© 2015 Schoenfelder et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Anguita E, Sharpe JA, Sloane-Stanley JA, Tufarelli C, Higgs DR, Wood WG. 2002. Deletion of the mouse α-globin regulatory element (HS –26) has an unexpectedly mild phenotype. Blood 100: 3450–3456. - PubMed
-
- Apostolou E, Thanos D. 2008. Virus infection induces NFKB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell 134: 85–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
