A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish
- PMID: 25752963
- PMCID: PMC4379706
- DOI: 10.1016/j.devcel.2015.01.032
A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish
Abstract
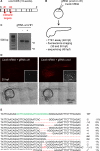
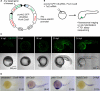
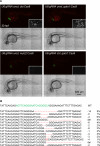
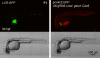
CRISPR/Cas9 technology of genome editing has greatly facilitated the targeted inactivation of genes in vitro and in vivo in a wide range of organisms. In zebrafish, it allows the rapid generation of knockout lines by simply injecting a guide RNA (gRNA) and Cas9 mRNA into one-cell stage embryos. Here, we report a simple and scalable CRISPR-based vector system for tissue-specific gene inactivation in zebrafish. As proof of principle, we used our vector with the gata1 promoter driving Cas9 expression to silence the urod gene, implicated in heme biosynthesis, specifically in the erythrocytic lineage. Urod targeting yielded red fluorescent erythrocytes in zebrafish embryos, recapitulating the phenotype observed in the yquem mutant. While F0 embryos displayed mosaic gene disruption, the phenotype appeared very penetrant in stable F1 fish. This vector system constitutes a unique tool to spatially control gene knockout and greatly broadens the scope of loss-of-function studies in zebrafish.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

References
-
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 OD017870/OD/NIH HHS/United States
- R01 DK53298/DK/NIDDK NIH HHS/United States
- P30 DK049216/DK/NIDDK NIH HHS/United States
- P01 HL032262/HL/NHLBI NIH HHS/United States
- P30 DK49216/DK/NIDDK NIH HHS/United States
- R01 HL048801/HL/NHLBI NIH HHS/United States
- R01 HL04880/HL/NHLBI NIH HHS/United States
- P01 HL32262/HL/NHLBI NIH HHS/United States
- R01 DK053298/DK/NIDDK NIH HHS/United States
- U01 HL10001/HL/NHLBI NIH HHS/United States
- R01 CA103846/CA/NCI NIH HHS/United States
- U01 HL100001/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R24 DK092760/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

