Organization of TNIK in dendritic spines
- PMID: 25753355
- PMCID: PMC4509985
- DOI: 10.1002/cne.23770
Organization of TNIK in dendritic spines
Abstract
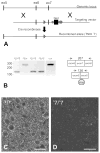
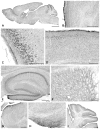
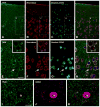
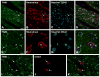
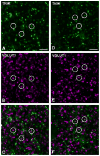
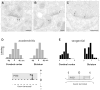
Tumor necrosis factor receptor-associated factor 2 (TRAF2)- and noncatalytic region of tyrosine kinase (NCK)-interacting kinase (TNIK) has been identified as an interactor in the psychiatric risk factor, Disrupted in Schizophrenia 1 (DISC1). As a step toward deciphering its function in the brain, we performed high-resolution light and electron microscopic immunocytochemistry. We demonstrate here that TNIK is expressed in neurons throughout the adult mouse brain. In striatum and cerebral cortex, TNIK concentrates in dendritic spines, especially in the vicinity of the lateral edge of the synapse. Thus, TNIK is highly enriched at a microdomain critical for glutamatergic signaling.
Keywords: PSD; RRID:AB_11213019; RRID:AB_1858225; RRID:AB_94396; RRID:nif-0000-30467; TNIK; cerebral cortex; dendritic spine; striatum RRID:AB_11212843.
© 2015 Wiley Periodicals, Inc.
Conflict of interest statement
BB, QW, MDE, ML, GF, QL, NT are/were employees and/or shareholders of Pfizer, Inc. The other authors verify that they have no known or potential conflict of interest including any financial, personal, or other relationships with other people or organizations within 3 years of beginning the submitted work that could inappropriately influence, or be perceived to influence, this work.
Figures
References
-
- Armstrong DM, Saper CB, Levey AI, Wainer BH, Terry RD. Distribution of cholinergic neurons in rat brain: demonstrated by the immunocytochemical localization of choline acetyltransferase. J Comp Neurol. 1983;216(1):53–68. - PubMed
-
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O’Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Molecular psychiatry. 2012;17(9):887–905. - PMC - PubMed
-
- Bernard V, Bolam JP. Subcellular and subsynaptic distribution of the NR1 subunit of the NMDA receptor in the neostriatum and globus pallidus of the rat: co-localization at synapses with the GluR2/3 subunit of the AMPA receptor. Eur J Neurosci. 1998;10(12):3721–3736. - PubMed
-
- Brigidi GS, Bamji SX. Cadherin-catenin adhesion complexes at the synapse. Current opinion in neurobiology. 2011;21(2):208–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

