ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis
- PMID: 25753424
- PMCID: PMC7405934
- DOI: 10.1016/j.celrep.2015.02.014
ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis
Abstract
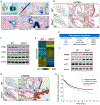
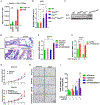
Lung cancer remains the leading cause of cancer death. Genome sequencing of lung tumors from patients with squamous cell carcinoma has identified SMAD4 to be frequently mutated. Here, we use a mouse model to determine the molecular mechanisms by which Smad4 loss leads to lung cancer progression. Mice with ablation of Pten and Smad4 in airway epithelium develop metastatic adenosquamous tumors. Comparative transcriptomic and in vivo cistromic analyses determine that loss of PTEN and SMAD4 results in ELF3 and ErbB2 pathway activation due to decreased expression of ERRFI1, a negative regulator of ERBB2 in mouse and human cells. The combinatorial inhibition of ErbB2 and Akt signaling attenuate tumor progression and cell invasion, respectively. Expression profile analysis of human lung tumors substantiated the importance of the ErbB2/Akt/ELF3 signaling pathway as both a prognostic biomarker and a therapeutic drug target for treating lung cancer.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Anastasi S, Sala G, Huiping C, Caprini E, Russo G, Iacovelli S, Lucini F, Ingvarsson S, and Segatto O (2005). Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene 24, 4540–4548. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

