Bactericidal efficiency and modes of action of the novel antimicrobial peptide T9W against Pseudomonas aeruginosa
- PMID: 25753629
- PMCID: PMC4432164
- DOI: 10.1128/AAC.04830-14
Bactericidal efficiency and modes of action of the novel antimicrobial peptide T9W against Pseudomonas aeruginosa
Abstract
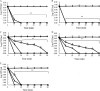
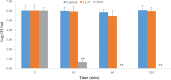
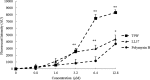
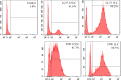
The antipseudomonal efficiency and mechanism of action of a novel engineered antimicrobial peptide, T9W, were evaluated in this study. T9W displayed high activity, with a lethal concentration (LC) of 1 to 4 μM against Pseudomonas aeruginosa, including against ciprofloxacin-, gentamicin-, and ceftazidime-resistant strains, even in the presence of 50 to 300 mM NaCl, 1 to 5 mM Ca(2+), or 0.5 to 2 mM Mg(2+). The time-kill curve (TKC) analysis demonstrated concentration-dependent activity, with T9W achieving complete killing in less than 30 min at 1× LC and in less than 5 min at 4× LC. Combination TKC analyses additionally demonstrated a synergistic effect with ciprofloxacin and gentamicin. The selectivity of T9W was further supported by its ability to specifically eliminate P. aeruginosa in a coculture with macrophages without toxicity to the mammalian cells. The results from fluorescent measurement indicated that T9W bound to lipopolysaccharide (LPS) and induced P. aeruginosa membrane depolarization, and microscopic observations and flow cytometry further indicated that T9W targeted the P. aeruginosa cell membrane and disrupted cytoplasmic membrane integrity, thereby causing cellular content release leading to cell death. This study revealed the potential usefulness of T9W as a novel antimicrobial agent against P. aeruginosa.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
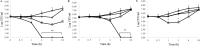
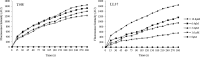


References
-
- Strateva T, Mitov I. 2011. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann Microbiol 61:717–732. doi:10.1007/s13213-011-0273-y. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

