The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling
- PMID: 25753661
- PMCID: PMC4381082
- DOI: 10.1093/nar/gkv196
The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling
Abstract
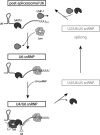
The spliceosome is a large molecular machine that serves to remove the intervening sequences that are present in most eukaryotic pre-mRNAs. At its core are five small nuclear ribonucleoprotein complexes, the U1, U2, U4, U5 and U6 snRNPs, which undergo dynamic rearrangements during splicing. Their reutilization for subsequent rounds of splicing requires reversion to their original configurations, but little is known about this process. Here, we show that ZK863.4/USIP-1 (U Six snRNA-Interacting Protein-1) is a ribonucleotidyl transferase that promotes accumulation of the Caenorhabditis elegans U6 snRNA. Endogenous USIP-1-U6 snRNA complexes lack the Lsm proteins that constitute the protein core of the U6 snRNP, but contain the U6 snRNP recycling factor SART3/B0035.12. Furthermore, co-immunoprecipitation experiments suggest that SART3 but not USIP-1 occurs also in a separate complex containing both the U4 and U6 snRNPs. Based on this evidence, genetic interaction between usip-1 and sart-3, and the apparent dissociation of Lsm proteins from the U6 snRNA during spliceosome activation, we propose that USIP-1 functions upstream of SART3 to promote U6 snRNA recycling.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
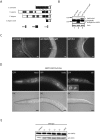

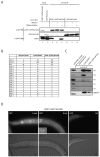

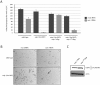
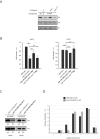
References
-
- Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell. Mol. Biol. 2009;272:149–189. - PubMed
-
- Staley J.P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. - PubMed
-
- Vijayraghavan U., Company M., Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. - PubMed
-
- Shannon K.W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

