Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7
- PMID: 25753663
- PMCID: PMC4381077
- DOI: 10.1093/nar/gkv173
Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7
Abstract
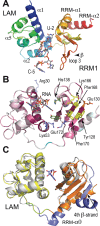
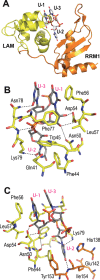
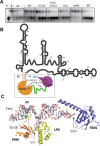
The non-coding RNA 7SK is the scaffold for a small nuclear ribonucleoprotein (7SKsnRNP) which regulates the function of the positive transcription elongation factor P-TEFb in the control of RNA polymerase II elongation in metazoans. The La-related protein LARP7 is a component of the 7SKsnRNP required for stability and function of the RNA. To address the function of LARP7 we determined the crystal structure of its La module, which binds a stretch of uridines at the 3'-end of 7SK. The structure shows that the penultimate uridine is tethered by the two domains, the La-motif and the RNA-recognition motif (RRM1), and reveals that the RRM1 is significantly smaller and more exposed than in the La protein. Sequence analysis suggests that this impacts interaction with 7SK. Binding assays, footprinting and small-angle scattering experiments show that a second RRM domain located at the C-terminus binds the apical loop of the 3' hairpin of 7SK, while the N-terminal domains bind at its foot. Our results suggest that LARP7 uses both its N- and C-terminal domains to stabilize 7SK in a closed structure, which forms by joining conserved sequences at the 5'-end with the foot of the 3' hairpin and has thus functional implications.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Maris C., Dominguez C., Allain F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. - PubMed
-
- Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

